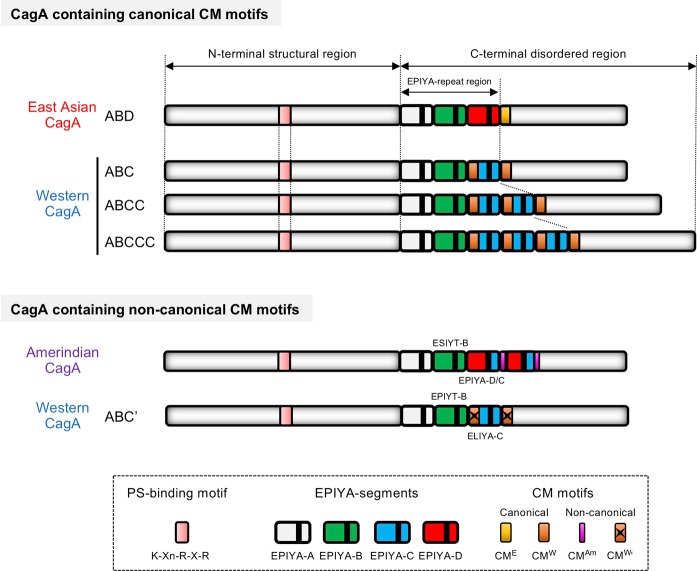
Fig. 1. Schematic structure of the H. pylori CagA protein.
The CagA protein is composed of a structural N-terminal region and an intrinsically disordered C-terminal region. The K-Xn-R-X-R motif is required for CagA to physically associate with the membrane phospholipid phosphatidylserine (PS) in cells. The EPIYA (Glu-Pro-Ile-Tyr-Ala) motifs in the C-terminal region are the tyrosine phosphorylation sites of CagA. The EPIYA-repeat region of CagA includes the common EPIYA segments, EPIYA-A, EPIYA-B, and an East Asian CagA-specific EPIYA-D segment, or a variable number of Western CagA-specific EPIYA-C segments, which are a results of the sequence polymorphisms of the CagA protein. The CM motif, which is composed of 16 amino acid residues, serves as a PAR1b binding site, promoting the multimerization of the CagA protein. Based on sequence polymorphisms, the CM motif is subdivided into 2 groups: a canonical CM motif, which has conserved PAR1b-binding ability, and a noncanonical CM motif, which lacks the binding ability. East Asian CagA possesses a single East Asian CagA-specific CM motif (CME), whereas Western CagA possesses multiple Western CagA-specific CM motifs (CMW). The structure of CagA with noncanonical CM motifs, which lack binding ability to PAR1b/MARK2, is shown (bottom panel). The Amerindian CagA, which is derived from the H. pylori v225 strain, possesses an internally deleted noncanonical CM motif. The ABC’-type Western CagA, which was cloned from the H. pylori TH2099 strain that had colonized housed macaques, possesses a derivative of the CMW motif with amino acid substitutions (CMW’) as well as the atypical EPIYA-C segment that contains the ELIYA sequence.