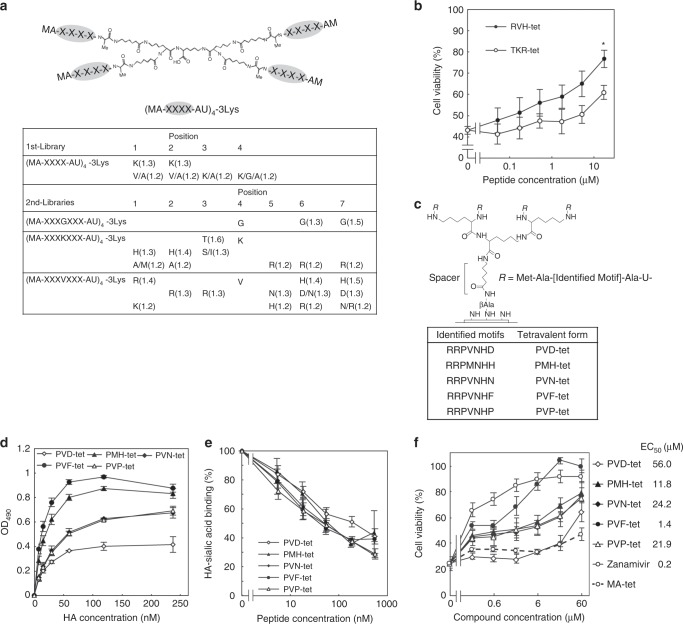
Fig. 1. Multivalent peptide library screen identified peptide-based HA inhibitors.
a The tetravalent peptide library for the first screen comprised tetravalent compounds with four randomized peptides of sequence Met-Ala-X-X-X-X-Ala-U (U; amino hexanoic acid), where X indicates any amino-acid except Cys. The library was screened to identify compounds that bound to HA but not to HA L194A. Three similar libraries with randomized peptides of sequence Met-Ala-X-X-X-(Gly/Lys/Val)-X-X-X-Ala-U were used for the second round of selections. Values in parentheses indicate the relative selectivity of the compound with indicated amino acids at the indicated positions. Each screen was performed twice; representative values are shown. b Two tetravalent peptides reduced IAV cytopathicity in MDCK cells. MDCK cells were infected with IAV strain PR8 at 10 MOI for 24 h in the presence of each peptide. Data are presented as a percentage of the control value without infection. *P < 0.05 (compared with TKR-tet by two-sided Student’s t test). c Structure of the tetravalent peptides synthesized on a cellulose membrane and optimized to HA. The density of the tetravalent peptide was set to 100% by using Fmoc-β-Ala-OH without Boc-β-Ala-OH for the first peptide synthesis cycle, and the number of amino hexanoic acid residues (U; spacer length) was set to one. Five HA-binding motifs identified by two steps of affinity maturation screening of the membranes are shown (lower panel). d ELISA to measure the binding of each tetravalent peptide (2 μm) to recombinant HA. e AlphaScreen assay to examine the inhibitory effect of tetravalent peptides on the binding between HA and its receptor mimic, α2–3-sialyllactose-PAA. Data are presented as a percentage of the control value. f The effects of tetravalent peptides and zanamivir on the IAV cytopathicity. MDCK cells were incubated with each compound for 30 min, and then infected with IAV strain PR8 at 10 MOI for 24 h. Data are presented as a percentage of the control value without infection. All data are from three independent experiments b and d–f, mean ± SEM). Source data are provided as a Source Data file.