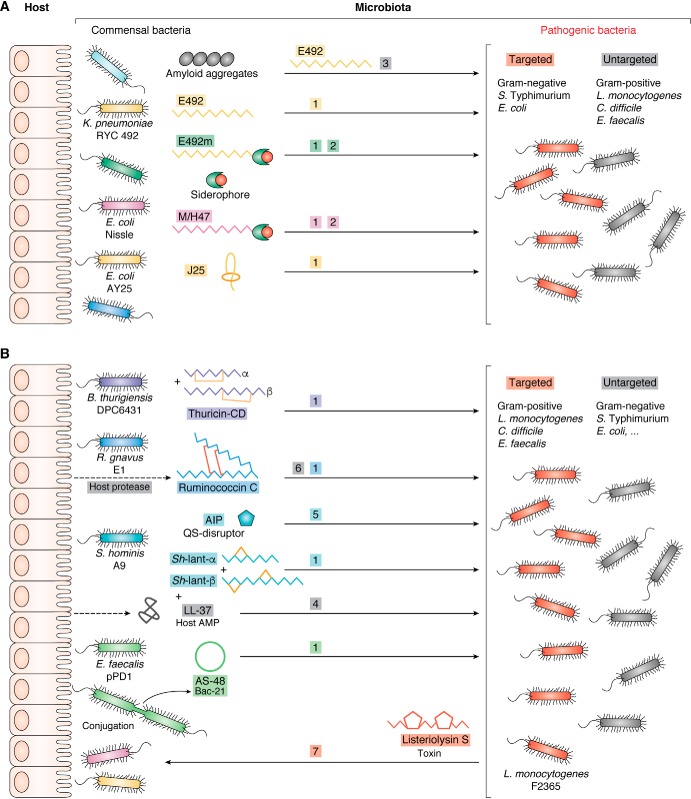
Figure 3.
Schematic of the main strategies of Gram-negative (A) and -positive (B) bacteria for colonization resistance and competition involving RPNPs. 1, interference competition using RPNPs as contact-dependent killer molecules (several examples from both Gram-negative and -positive bacteria, among which are the lasso peptide microcin J25 and the two-peptide sactibiotic thuricin CD, respectively); 2, exploitative competition exemplified by the battle for iron involving competition between siderophores and siderophore-modified peptides for the same high affinity receptors (MccE492m from K. pneumoniae RYC 492 and MccH47 from E. coli Nissle 1917); 3, reservoir of killing RPNPs using functional amyloids for delivering killers (MccE492 from K. pneumoniae RYC 492); 4, synergy between RPNPs and host AMPs (Sh-lantibiotic-α and Sh-lantibiotic-β from S. hominis A9, each synergizing with the host cathelicidin LL-37); 5, quorum-sensing interference (an autoinducing RPNP from S. hominis A9 acts as a disruptor of quorum sensing that inhibits the secretion of the virulence factor PSMα in S. aureus; 6, cooperation between host and RPNP producing bacterium in the gut for the final processing step leading to mature RPNP acting by interference competition (ruminococcin C from R. gnavus E1); 7, manipulation of commensals by the pathogen to occupy the niche and increase virulence (the pathogenic strain L. monocytogenes F2365 produces listeriolysin S, a TOMM virulence factor, which targets commensal bacteria occupying the same niche and devoid of self-immunity to the toxin, to increase nutrient level and favor invasion).