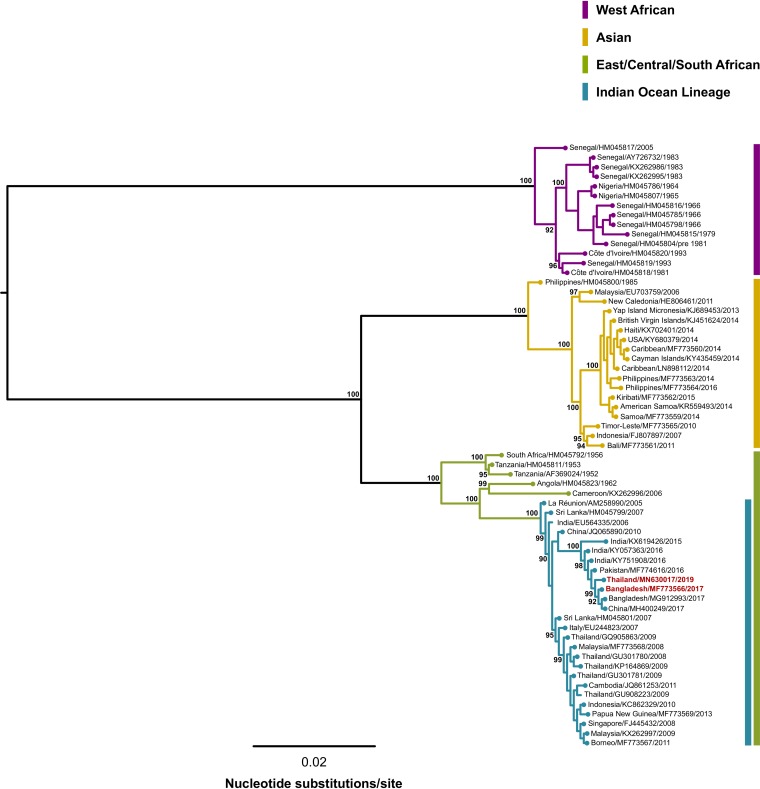
FIG 1.
Approximately maximum-likelihood phylogenetic tree (midpoint rooted for branch visibility only) inferred for 63 CHIKV genome sequences (excluding 5′ and 3′ untranslated regions) using FastTree version 2.1.11 software and the GTR nucleotide substitution model with the default setting of 20 for rate categories of sites (8). Percentage Shimodaira-Hasegawa-like local support values are shown for key nodes. Multiple sequence alignments were performed using the Multiple Alignment using Fast Fourier Transform (MAFFT) program version 7.450 and Geneious version 10.2.6 software. The three major CHIKV genotypes (West African, Asian, and ECSA) are shown, including grouping of the Bangladesh 2017 (GenBank accession number MF773566) and Thail 2019 (GenBank accession number MN630017) isolates (red font) within the ECSA-Indian Ocean lineage.