Abstract
Schizophrenia is a debilitating psychiatric disorder with a complex genetic architecture and limited understanding of its neuropathology, reflected by the lack of diagnostic measures and effective pharmacological treatments. Geneticists have recently identified more than 145 risk loci comprising hundreds of common variants of small effect sizes, most of which lie in noncoding genomic regions. This review will discuss how the epigenetic toolbox can be applied to contextualize genetic findings in schizophrenia. Progress in next-generation sequencing, along with increasing methodological complexity, has led to the compilation of genome-wide maps of DNA methylation, histone modifications, RNA expression, and more. Integration of chromatin conformation datasets is one of the latest efforts in deciphering schizophrenia risk, allowing the identification of genes in contact with regulatory variants across 100s of kilobases. Large-scale multiomics studies will facilitate the prioritization of putative causal risk variants and gene networks that contribute to schizophrenia etiology, informing clinical diagnostics and treatment downstream.
Keywords: schizophrenia, epigenetics, higher-order chromatin, histone modification, multiomics, GWAS
Abstract
La esquizofrenia es un trastorno psiquiátrico invalidante con una arquitectura genética compleja y una comprensión limitada de su neuropatología, lo que se traduce en la ausencia de mediciones diagnósticas y tratamientos farmacológicos eficaces. Recientemente los genetistas han identificado más de 145 loci de riesgo que comprenden cientos de variantes comunes de pequeño efecto, la mayoría de los cuales se encuentran en regiones genómicas no codificantes. Este artículo revisará cómo se pueden aplicar las herramientas epigenéticas para contextualizar los hallazgos genéticos en la esquizofrenia. El progreso en la secuenciación de segunda generación, junto con la creciente complejidad metodológica, ha llevado a la recopilación de mapas de metilación de ADN, modificaciones de histonas, expresión de ARN y otros más. Uno de los últimos esfuerzos para descifrar el riesgo de presentar esquizofrenia es la integración de los conjuntos de datos de conformación de cromatina, lo que permite la identificación de genes en contacto con variantes reguladoras en cientos de kilobases. Los estudios multiómicos a gran escala facilitarán la priorización de las posibles variantes de riesgo causal y las redes de genes que contribuyen a la etiología de la esquizofrenia, lo que incidirá en el diagnóstico clínico y en el tratamiento.
Abstract
La schizophrénie est une maladie psychiatrique invalidante dont l’architecture génétique est complexe : sa neuropathologie est mal comprise comme le montre le manque de mesures diagnostiques et de traitements médicamenteux efficaces. Les généticiens ont récemment identifié plus de 145 loci de risque comprenant des centaines de variants courants de faible effet, la plupart situés dans des régions non codantes du génome. Cet article discute de la façon d’appliquer les outils épigénétiques pour replacer les résultats génétiques dans le contexte de la schizophrénie. Les avancées du séquençage de nouvelle génération associées à une méthodologie de plus en plus complexe, ont permis de regrouper les cartes pangénomiques de la méthylation de l’ADN, des modifications de l’histone, de l’expression de l’ARN et plus encore. L’un des derniers progrès en date dans le déchiffrage du risque schizophrénique porte sur l’intégration de séries de données sur l’architecture de la chromatine, permettant l’identification de gènes en contact avec les variants de régulation parmi des centaines de kilobases. Des études multiomiques à grande échelle aideront à prioriser les variants du risque causal putatif et les réseaux de gènes qui contribuent à l’étiologie de la schizophrénie, pour renseigner en aval les diagnostics cliniques et les traitements.
Introduction
Schizophrenia (SZ) is a complex and debilitating neuropsychiatric illness that has approximately 1% average lifetime prevalence globally, although rates vary regionally up to fivefold. 1 - 3 The disease is characterized by “positive” symptoms, consisting of recurrent psychosis, hallucinations, and disorganized speech; “negative” symptoms, including anhedonia, social withdrawal, and flattened affect; and broad cognitive dysfunction. 1 Devastatingly, those diagnosed with SZ have a staggeringly high unemployment rate of 80% to 90% 4 , 5 and a life expectancy that is reduced by 10 to 20 years, with mortality often caused by suicide or cardiovascular conditions. 2 , 6 An incredibly isolating disease on a personal level, SZ also poses a great economic burden in terms of health and social care. 7
SZ is thought to result from an intricate choreography of genetic and environmental factors throughout early brain development that predispose an individual to the disease. Heritability estimates range up to 80%, underlining the importance of genetic contributions to the calculation of SZ disease risk. 7 - 9 The field has benefited greatly from the advent of next-generation sequencing and large-scale genome-wide association studies (GWAS) that have identified a combination of rare and common variants that are associated with SZ risk, spurring on the investigation of potential etiological mechanisms. Evidence from both genetic and epidemiological studies point to a disease of impaired early, potentially prenatal, brain development. 10
A disease with heterogeneous clinical presentation, SZ’s non-Mendelian genetic risk architecture is equally heterogeneous and nuanced, with contributions from rare penetrant variants, such as copy number variants (ie, deletions and duplications of chromosomal regions) to common variants of low to modest effect size. 11 The most recent SZ genome-wide association study (GWAS), performing a meta-analysis of approximately 41 000 cases and 65 000 controls, identified 145 risk loci, composed of a collection of single nucleotide polymorphisms (SNPs) in linkage disequilibrium (LD), adding 50 more loci to those from an earlier study. 12 , 13 However, despite advances in mapping the complex genetic risk architecture of SZ, successful drug development has still been elusive due to lack of a unifying neuropathology and difficulties in identifying the likely casual genes and vulnerable cellular contexts. In this regard, neuroepigenetics—broadly defined as the study of chromatin configuration and function from development to adulthood in the nervous system—could provide an important perspective in prioritizing risk variants and their target genes, paving the way for novel diagnostic and therapeutic approaches. 14
The beginnings of epigenetic mechanisms in schizophrenia
Early work in SZ epigenetics included investigating DNA cytosine methylation of candidate gene promoters such as RELN (glycoprotein for neurodevelopment and migration), 15 , 16 GAD1 (glutamic acid decarboxylase 1), 17 COMT (catechyl-O-methyltransferase), 18 , 19 and SOX10 (developmental transcription factor) 19 , 20 to name a few examples. Specifically, the promoters of RELN and GAD1 , both involved in the γ-aminobutyric acid (GABA) pathway important for inhibitory neurotransmission, are hypermethylated in SZ, 15 - 17 which is consistent with reduced expression of the two genes in postmortem brain of SZ patients. 21 - 23 Likewise, the promoter of oligodendrocyte-specific transcription factor SOX10 is hypermethylated in patients with SZ, in concordance with its downregulation in the diseased brain as well. 20 On the other hand, the promoter of COMT , a dopaminergic pathway component, is hypomethylated in postmortem SZ brains. 19 While the early studies were illustrative in tying together potential epigenetic mechanisms and expression patterns for important neurobiological genes, they did not allow for scalable exploration of the whole genome and required knowledge of candidate genes to design the study. More recent studies using techniques that allow genome-wide interrogation, through high-throughput next-generation sequencing, are able to substantially increase sample sizes, finding many SZ-associated differentially methylated regions distributed throughout the genome in blood (689 cases, 645 controls) 24 and across four different brain regions (41 cases, 46 controls). 25 To dissect the association between methylation and SZ risk further, one study combined high-density methylation profiling with genome-wide SNP genotyping in 166 human fetal brain samples (56 to 166 days post-conception) to find >16 000 methylation quantitative trait loci (mQTLs), instances of DNA methylation that can be influenced by variation in the sequence. 25 Moreover, these fetal brain mQTLs, many of which are remarkably stable even in the adult brain, were significantly enriched amongst SZ risk loci, allowing for the homing in on discrete sites of methylation in the fetal brain that also harbor SZ risk variants. 25
Histone post-translational modifications (PTMs) are another layer of epigenetic control that contribute to higher-order chromatin regulation, such that combinatorial PTMs establish various chromatin states, including active or silenced transcription, regulatory sequences, etc. 26 Like in early methylation studies of SZ, histone PTMs were assayed in single-gene fashion. 27 For instance at the GAD1 locus, Huang et al discovered decreased histone 3 lysine 4 trimethylation (H3K4me3), associated with the transcriptional process, in the prefrontal cortex (PFC) of predominantly female patients with SZ when compared with controls at the 5’ end of the GAD1 gene, overlapping with a SZ risk locus. 28 A later study detected decreased H3K9K14 acetylation, also marking transcriptionally active chromatin, at several candidate genes, including GAD1 . 29 Eventually, the chromatin immunoprecipitation (ChIP) assay used in these two studies was refined and made scalable to high-throughput library generation in postmortem brain, taking on the form of ChIP with next-generation sequencing (ChIP-Seq) to query the whole genome, and not just at predetermined sites, in a cell type-specific fashion. 30 Only recently has this approach been applied to the study of psychiatric disorders, specifically SZ. A study of 157 open chromatin-associated histone profile (H3K4me3 and H3K27ac) reference maps from PFC and anterior cingulate cortex (ACC) of sorted neuronal (NeuN+) and non-neuronal (NeuN-) cell populations revealed differing landscapes at loci based on cell type and brain region. 31 Notably, the authors found a striking overrepresentation of risk variants for SZ that was highly specific to neuronal chromatin, as determined by partitioning heritability, highlighting the need to study epigenetic contributions to disease in a cell type-specific fashion. 31 This is especially important when considering the fact that around half of the noncoding regions could have regulatory functions specific to a given tissue, cell type, or developmental stage. 32 , 33 In the brain, this nuance is compounded by the enormous cellular heterogeneity and long developmental trajectories that occurs to a lesser degree in other tissues. 34
Given that the majority of SZ risk variants occur in these noncoding (ie, intronic or intergenic) regions of the genome, it is difficult to identify the associated causal gene, when considering that the variants could be in regulatory elements, such as enhancers or repressors, impacting the expression of distally located genes. 13 , 35 Indeed, using heritability partitioning, the authors found that 16% of imputed SNPs overlapping DNAse hypersensitivity sites—indicators of open chromatin and regulatory regions—explained an average of 79% of the SNP heritability spanning 11 diseases, including SZ. 36 One approach is to leverage RNA-sequencing (RNA-seq) data with SNPs to identify genetic variants that are correlated with expression of genes, even if they are not immediately adjacent, called expression quantitative trait loci (eQTL). This is precisely what the CommonMind Consortium did, using expression data from dorsolateral PFC of people with SZ (N=258) and control subjects (N =279) 37 and imputed SNP genotypes. They found that, of the 108 loci previously reported, 13 20 of them (~20%) had eQTL SNP-gene pairs within 1 megabase (Mb) of linear genomic distance (ie, cis eQTLs) that might contribute to altered gene expression and SZ liability. 37 A recent study was able to expand the number of GWAS loci colocalized with eQTL signal from 20 to 40 by including conditional eQTLs (ie, not just one primary eQTL for each gene), beginning to consider epigenetic and cell states within which different SNPs might function. 38
Assay for transposase-accessible chromatin with sequencing (ATAC-seq) also offers glimpses into open chromatin regions (OCRs) along with footprints of DNA-binding factors, such as transcription factors. 39 One study of sorted neuronal and non-neuronal cells from postmortem PFC identified cell type-specific OCRs, with neuronal OCRs more enriched for distal regulatory elements and evolutionarily conserved regions, as well as a subset of transcription factors highly enriched in SZ loci specific to neurons, providing a potential functional role. 40 An expanded study assayed the same two cell types and 14 different brain regions in five individuals, reporting higher regional variability in neuronal chromatin and significant enrichment only for neuropsychiatric traits, including SZ, in cell- and region-specific fashion, 41 again highlighting the need for approaches that take into account the many nuances of the brain. A case-control study of chromatin accessibility (142 SZ, 143 control) only identified three differentially accessible regions, suggesting that differences could be subtle between diseases and healthy PFC, 42 although looking at cell type-specific maps may have improved the results. Taken together, such GWAS-guided ChIP-seq, eQTL, and ATAC-seq studies have begun to shed light on possible mechanisms in which SNPs located in noncoding portions of the human genome may exert a regulatory influence on gene expression, even distally, if they happen to coincide with functional elements such as enhancers or repressors.
Recently, the field has enjoyed a wave of “post-GWAS” analyses aimed at increasing the power to prioritize causal genes by performing gene-based, as opposed to single-variant-based, associations, with the underlying assumption that many genetic variants influence traits, like SZ, via transcriptional regulation. 43 By focusing on the genetic component of expression, environmental factors are excluded from consideration, thereby increasing statistical power. 44 One of these methods, called the transcriptome-wide association study (TWAS), 45 has revealed 157 significant genes, 35 of which did not overlap a known GWAS locus. 46 Of 157 genes, 42 genes were associated with specific chromatin phenotypes (ie, histone modification ChIP-seq peaks), suggesting putative regulatory mechanisms. Moreover, 105/157 TWAS-predicted risk genes overlapped with genes linked to index SNPs through chromatin interaction data from fetal brain. 46 The method also revealed that the TWAS genes were more significantly associated with the trait in question than the nearest gene and had stronger eQTL effects at the index SNP, 47 emphasizing the need to go beyond the linear genome. A similar method called PrediXcan 48 has been applied to the largest transcriptomic imputation study in SZ to date, identifying 67 genic associations across 13 brain regions, 19 of which were novel. 49 These genes were involved in new biological pathways such as hexaminidase-A and porphyrin metabolism, both of which have historic connections to SZ without much prior genetic evidence supporting them. 49 Recently, this technique has again confirmed that most trait associations are tissue-specific, underscoring the necessity of studying homogeneous populations of specific cell types. 50
Large-scale functional genomic and epigenetic studies
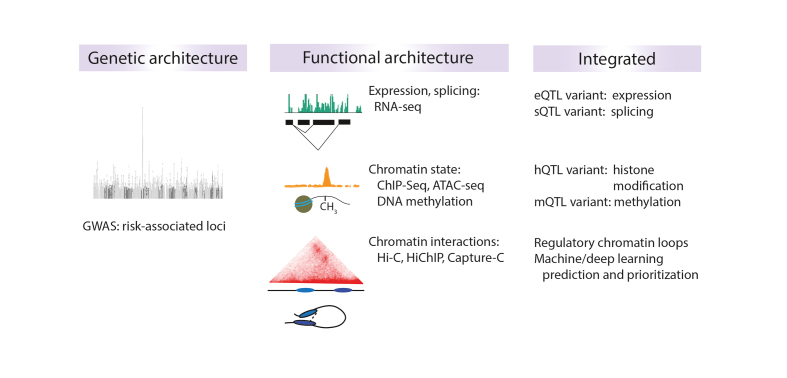
Looking ahead, the most promising approaches could involve integration of multiple orthogonal datasets taking into account cell type contexts, particularly with larger sample sizes, in order to triangulate more functionally implicated genetic variants associated with SZ ( Figure 1 ). Leveraging both RNA-seq and ATAC-seq data, one study identified 118 differentially transcribed enhancer RNAs (eRNAs), important for activity-dependent gene regulation in the human brain, in SZ compared with controls. The same study also found genetic variants that affect expression of approximately 1000 enhancers. 51 A recent multiomics study of tissue and single- cell RNA-seq, histone modification landscapes, CTCF transcription factor binding sites, DNA methylation, and genotypes (1230 samples from 48 brains; 18 288 single cells/nuclei from 12 brains) aimed to comprehensively chart the human neurodevelopmental trajectory, from the embryonic stage to adulthood, of gene expression programs and their regulation in various brain regions and cellular contexts. 52 With this integrative approach, the authors were able to identify a global transition in the transcriptomic profile during late fetal development where there was a sharp reduction in differences across brain regions coincident with increased signatures of expression of genes important for neuronal structure and activity. 52 Importantly, using partitioned linkage disequilibrium score regression analysis, they found that SZ SNP heritability was enriched in the dorsolateral PFC-specific regulatory elements as identified by H3K27ac peaks. With regard to cell type context, cortical excitatory neurons, embryonic/fetal progenitor cells, and adult cortical interneurons were enriched for expression of high-stringency genes associated with SZ, 52 echoing another single-cell study that found similar cellular patterns. 53 Analysis of gene coexpression modules revealed one particular module enriched for genes enriched in fetal and adult excitatory neurons associated with SZ, fetal enhancers, neuronal (as opposed to neural progenitor or glial) expression, and neuronal undermethylated sites. 52 This module, consisting of 145 total genes, included MEF2C and SATB2 , which have previously been associated with neurodevelopmental disorders in general and specifically SZ. 13 , 54 - 56
Figure 1. Advancing genetic findings with functional epigenomic information and integration. Genetic analyses identify common variation associated with schizophrenia (SZ) risk using genome-wide association studies (GWAS), as indicated by the Manhattan plot (left). Most variants do not alter protein structure and may have diverse regulatory functions (eg, enhancer, repressor, regulator of splicing, etc). Quantitative mapping approaches (middle) can measure transcript abundance and splicing (RNA-seq); chromatin state through histone modifications (ChIP-seq), DNA methylation, identification of active/ open chromatin (ATAC-seq, DNase-seq); and higher-order chromatin conformations (Hi-C, HiChIP, Capture-C). Integration of these functional datasets with the genetic architecture from GWAS can identify putative causal variants that impact any one (or more) of the epigenetic features listed above, providing a mechanism for potential gene dysregulation converging on important neural and developmental pathways.
Another large study (over 2000 postmortem brain samples from individuals with SZ, bipolar disorder, and autism spectrum disorder as well as controls) looking at the transcript isoform level across SZ, bipolar disorder, and autism spectrum disorders, uncovered disease-specific differential splicing and expression and that isoform-level changes, as opposed to gene-level, showed the largest effect sizes and greatest disease specificity. 57 Of note, approximately 700 noncoding RNAs (including 208 long intergenic noncoding RNAs, or lincRNAs), which are thought to have a transcriptional regulatory role, were differentially expressed in SZ, specifically in excitatory neurons. 57 A TWAS performed on these data identified a stringent list of 64 significant genes, consistently prioritized by multiple methods, including downregulated lysine methyltransferases SETD6 and SETD8 along with brain-enriched lincRNA LINC00634 , 57 highlighting transcriptional and epigenetic regulation as important molecular mechanisms to consider in SZ risk. Taken together, these two studies testify to the power gained by leveraging large brain datasets across multiple modalities in order to identify the contributions of cell type-specific pathways and their regulation over time. Such “four-dimensional (4D) mapping,” including spatial and temporal components, will be vital to distinguish between the effects of direct genetic insult and its resulting indirect cascade of molecular events, as well as to identify when specifically in development intervention would be most ameliorative.
Schizophrenia risk-associated genome in 3D
Even with the identification of transcriptomic patterns and gene coexpression modules or the annotation of noncoding variants in the context of SZ risk, led in large part by the generation of brain-specific functional genomic datasets by the PsychENCODE consortium, 58 there is still a part of the story that is missing. We cannot simply assume that noncoding SZ variants, which make up most of the genetic risk architecture, act on the genes that are closest to them on the linear genome 59 , 60 or are in linkage disequilibrium. 61 Therefore, it is necessary to consider the genome in three dimensions and map chromatin interactions that can bring distal regions into close physical proximity, in many cases for fine-tuning gene expression through regulatory elements (eg, promoter-enhancer interactions ( Figure 1 )). 62
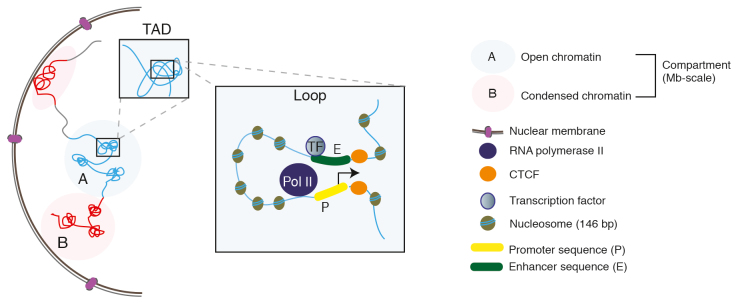
The three dimensional conformation of chromatin and entire chromosomes is referred to as the “3D genome” or 3DG. Chromosome conformation capture methods combined with deep sequencing, such as in situ Hi-C, are used to study the 3DG. 63 Briefly, the technique involves crosslinking cells or tissue, isolating intact nuclei, digesting the chromatin within the nuclei with restriction enzymes, biotinylating cut ends, ligating fragments in close proximity, shearing the DNA, pulling down biotinylated chimeric fragments, and generating libraries for deep paired-end sequencing. 63 Variants of the technique include capture Hi-C to isolate fragments of interest with baits, 64 , 65 single cell Hi-C, 66 - 68 Hi-C paired with ChIP (HiChIP), 69 and low input Hi-C, 70 - 72 among others. This cadre of methods has enumerated various principles of 3DG organization (see Figure 2 for more details). Perhaps the biggest layer of the 3DG involves the clustering of euchromatic (A) and heterochromatic (B) sequences into regions of approximately ~5 megabases (Mb) called “compartments” 73 ( Figure 2 ), although recent reports say that they may be at a smaller scale than previously envisioned (15-300kb), often segregated by shared transcriptional states and histone modification landscapes. 63 , 74 , 75
Figure 2. The genome in 3D. Chromatin, organized as arrays of the nucleosome (146 bp of DNA wrapped in 2.5 loops around the core histone octamer), is arranged into A (open, euchromatic) or B (closed, heterochromatic) compartments that can be megabases wide. Superimposed upon these structures are topologically associating domains (TADs), which are on average (median size) 185 kb, and can be within either A or B compartments. Sequences within TADs are much more likely to come into contact with each other than with loci from outside domains. TAD boundaries and chromatin loop formations (eg, promoter-enhancer loops) are often demarcated by CTCF (and additional proteins not shown here). Chromatin loops, or interactions, allow distal regulatory elements, like enhancers, to come into contact with gene promoters in order to regulate gene expression, often in cell type-specific fashion, with modulation of the RNA Polymerase II transcriptional machinery.
The next component of the 3DG is topologically associating domains (TADs), which are genomic sequences that preferentially interact with themselves as opposed to regions outside of their boundaries. 76 , 77 They act as insulators, preventing inappropriate interactions between elements such as promoters and enhancers, which may lead to abnormal levels of gene expression. 76 , 78 Furthermore, TADs are thought to be more conserved across cell types, and even species, than are compartments. 77 , 79 - 81 However, their insulation capacity and interconnections with other regions have been shown to be subject to cell type-specific changes, at least during lineage specification when primed perhaps by transcription factor binding. 82 Additionally, TAD insulation is highly correlated with transcription, such that new borders are able to form at developmentally regulated gene promoters, suggesting that novel TAD landscapes may arise with certain contexts. 83
The spatial genome also includes chromatin loops or interactions, commonly defined as distinct pairwise contacts that, in Hi-C maps, sharply stand out from the surrounding “linear” genome background, often linking regulatory elements with distal gene promoters in a cell type-specific fashion. 63 The anchors of many chromatin loops (65% to 92%) often have CTCF, which in this case acts as an architectural scaffolding protein, in an inward/convergent orientation at binding sites 63 , 77 , 81 , 84 that, when experimentally inverted, could in some cases affect normal patterns of gene expression. 85
Because the majority of functional elements in noncoding portions of the human genome, such as enhancers and repressors, are not contacting the nearest TSS but instead are interacting with genes located elsewhere on the chromosome, 86 it is unsurprising therefore that non-3DG based approaches relying on purely linear genomic distance, had only limited success in assigning specific target genes to risk loci. 34 The inability to catalog reliably gene targets of SZ risk variants understandably hindered the downstream efforts to identify networks contributory to disease etiology. eQTLs, as discussed earlier, began to consider the impacts of common variants on distal genes; however, these are still mostly restricted to SNP-gene associations within 1 Mb and are computationally derived without using actual measurements of the 3DG space. 37 , 87 In a pioneering study, Won and colleagues generated Hi-C chromosomal conformations from fetal brain and mapped gene targets for schizophrenia GWAS noncoding variants; many of these genes were involved in disease-relevant pathways such as neurogenesis or cholinergic signaling. 60 Another Hi-C study generating 3DG maps from fetal and adult human cortex to explore GWAS loci for multiple neuropsychiatric disorders and traits found that chromatin interactions explained ~73% of genes implicated by eQTLs (2,292/3,121); strikingly, there are 4101 genes discovered only through Hi-C that are otherwise unaccounted for when considering only location or eQTLs. 88 For SZ specifically, the authors found that genes implicated by location were enriched for neuronal and synaptic processes, which are certainly important in disease etiology, whereas as those genes implicated by functional chromatin interaction data were enriched for regulatory chromatin biology, perhaps pointing to more upstream dysregulation. 88 While such datasets have advanced our understanding of the genetic risk architecture of psychiatric disease, 10 , 60 3DG mapping from postmortem tissue lacks cell type-specific resolution and may not capture higher-order chromatin structures sensitive to the autolytic process. 89
Two recent studies addressed some of these technical shortcomings. First, Hi-C mapping of human induced pluripotent stem cell (hiPSC)-derived neural progenitor cells (NPCs) and their isogenically differentiated excitatory neurons and glia increased the number of transcribed genes associated with SZ GWAS loci by approximately 2- to 3-fold (total N expressed genes connected to or located within an SZ GWAS locus ranged from 201-386, depending on cell type). 90 Since neurons, together with NPCs, had the greatest number of cell type-specific interactions anchored in a risk locus (as compared with glia), one could conclude that the 3DG space corresponding to SZ risk disproportionately affects neurons, 90 echoing other studies that report a neuronal burden in the SZ genetic architecture. 31 , 52 , 53 , 57 Remarkably, the SZ-associated chromosomal connectome (ie, GWAS loci and their connected genes) specific to NPC or excitatory neurons was associated with coordinated gene expression “clusters” and protein-protein interactions, where one cluster strongly enriched for regulators of neuronal connectivity and synaptic plasticity, and another cluster for chromatin-associated proteins, including transcriptional regulators, 90 similar to the results of Giusti-Rodriguez et al. 88 Many interactions of interest between noncoding SZ variants and neurally relevant genes, such as pro-neuronal transcription factor ASCL1 or members of the clustered Protocadherin family, were functionally validated in NPCs with CRISPR epigenomic and genomic editing. 91 Likewise, an expanded genome space involving higher-order chromatin interactions anchored in schizophrenia risk loci, prioritized with orthogonal CTCF and histone modification ChIP-seq datasets, has also been described for cultured primary sensory neurons from the olfactory neuroepithelium, also pointing to neurogenesis as an important gene set. 92
Furthermore, a massive multiomics study by the PsychENCODE consortium analyzed the open chromatin landscape, transcriptional histone marks, and the transcriptome from altogether 2000 postmortem brains, including hundreds of cases diagnosed with schizophrenia, and combined these “linear genome” profiles with Hi-C data from fetal and adult reference brains. 93 The study mapped ~79 000 brain-active enhancers with their associated chromosomal contacts and TAD landscapes and identified a vast number of eQTLs and gene regulatory networks; perhaps most importantly, the investigators applied deep machine learning algorithms that, for the first time, were able to predict presence or absence of disease (ie, SZ) based on a subject’s brain transcriptome and chromatin profiles. 93 The study approached disease prediction at a probability level of ~75%, reflecting a significant advancement over more conventional genomic approaches predicting disease only marginally above chance (50%). 93 Very recently, another study presented an integrative risk gene selector (iRIGS), a computational framework to integrate multiomics data, that predicted high-confidence risk genes that account for significantly enriched heritability, are expressed in brain tissues (especially prenatal), and are enriched for targets of already approved drugs, providing new opportunities to repurpose existing drugs for SZ. 94
Conclusions
The field of SZ epigenetics has experienced a recent surge of discovery, in part facilitated by innovation in the technical capacities of epigenomic architecture mapping and high-throughput sequencing. As a field, we have been able to progress from single candidate gene studies to probing the entire human genome, both the protein-coding and the hitherto enigmatic noncoding regions, in unbiased fashion across numerous modalities and cell types. However, the polygenic nature of SZ with its nuanced network of common variant influence requires more investigation to decipher, through both large-scale consortium-led efforts as well as incisive functional and mechanistic studies on discrete, contributory pathways. The many studies enumerated here in the new wave of SZ epigenetics research utilized high-resolution genomic and epigenomic datasets from relevant tissue and cell types, taking into account the 3DG space and leveraging orthogonal approaches, in order to arrive at higher confidence gene sets and predictive capacity. In this vein, the polygenic risk score (PRS), a metric that summarizes the inherited common variant burden in an individual, is interesting from a clinical precision-medicine diagnostic perspective. While being in the top 10% of PRSs carries a greater than 10-fold increased risk of SZ, 13 there is still a substantial overlap in the distribution of PRS between cases and controls, such that many controls are in the top decile and many cases in the lowest. 34 Deep-learning algorithms such as those employed by Wang et al pave the way forward, combining different slices of information to construct a more holistic functional genomic picture that is missed when considering simply the genetic information, thereby enhancing disease prediction. Another benefit of compiling and integrating many brain datasets is the identification of high-fidelity risk variant-gene interactions with regulatory epigenomic landscapes. What genes are revealed as important in multiple approaches? Do they converge on pathways with pathogenic potential? Are there druggable targets for major regulators of these pathways or hubs? Questions like these can help focus the expeditions ahead as we try to understand the genetic, functional, and cellular/network architecture of SZ culminating in disease.
Acknowledgments
PR is supported by predoctoral Ruth L. Kirschstein fellowship 1F30MH113330. The authors have no conflicts of interest to disclose
Contributor Information
Prashanth Rajarajan, Graduate School of Biomedical Sciences; Department of Psychiatry; Friedman Brain Institute; Icahn School of Medicine at Mount Sinai, New York, NY, US..
Schahram Akbarian, Department of Psychiatry; Friedman Brain Institute; Icahn School of Medicine at Mount Sinai, New York, NY, US..
REFERENCES
- 1.Kahn RS, Sommer IE, Murray RM, et al Schizophrenia. Nat Rev Dis Prim. 2015;1:15067. doi: 10.1038/nrdp.2015.67. [DOI] [PubMed] [Google Scholar]
- 2.McGrath J, Saha S, Chant D, Welham J. Schizophrenia: a concise overview of incidence, prevalence, and mortality. Epidemiol Rev. 2008;30:67–76. doi: 10.1093/epirev/mxn001. [DOI] [PubMed] [Google Scholar]
- 3.Bhugra D. The global prevalence of schizophrenia. PLos Med. 2005;2(5):e151. doi: 10.1371/journal.pmed.0020151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kooyman I, Dean K, Harvey S, Walsh E. Outcomes of public concern in schizophrenia. Br J Psychiatry. 2007;50(Supp):S29–S36. doi: 10.1192/bjp.191.50.s29. [DOI] [PubMed] [Google Scholar]
- 5.Marwaha S, Johnson S. Schizophrenia and employment - a review. Soc Psychiatry Psychiatr Epidemiol. 2004;39(5):337–349. doi: 10.1007/s00127-004-0762-4. [DOI] [PubMed] [Google Scholar]
- 6.Chesney E, Goodwin GM, Fazel S. Risks of all-cause and suicide mortality in mental disorders: a meta-review. World Psychiatry. 2014;13(2):153–160. doi: 10.1002/wps.20128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Owen MJ, Sawa A, Mortensen PB. Schizophrenia. Lancet. 2016;388(10039):86–97. doi: 10.1016/S0140-6736(15)01121-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry. 2003;60(12):1187–1192. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- 9.Hilker R, Helenius D, Fagerlund B, et al Heritability of schizophrenia and schizophrenia spectrum based on the nationwide Danish Twin Register. Biol Psychiatry. 2018;83(6):492–498. doi: 10.1016/j.biopsych.2017.08.017. [DOI] [PubMed] [Google Scholar]
- 10.de la Torre-Ubieta L, Stein JL, Won H, et al The dynamic landscape of open chromatin during human cortical neurogenesis. Cell. 2018;172(1-2):289–304. doi: 10.1016/j.cell.2017.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gershon ES, Grennan KS. Genetic and genomic analyses as a basis for new diagnostic nosologies. Dialogues Clin Neurosci. 2015;17(1):69–78. doi: 10.31887/DCNS.2015.17.1/egershon. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pardinas AF, Holmans P, Pocklington AJ, et al Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet. 2019;51(7):1193. doi: 10.1038/s41588-019-0450-7. [DOI] [PubMed] [Google Scholar]
- 13.Schizophrenia Working Group of the Psychiatric Genomics C Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511(7510):421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jakovcevski M, Akbarian S. Epigenetic mechanisms in neurological disease. Nat Med. 2012;18(8):1194–1204. doi: 10.1038/nm.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abdolmaleky HM, Cheng KH, Russo A, et al Hypermethylation of the reelin (RELN) promoter in the brain of schizophrenic patients: a preliminary report. Am J Med Genet B Neuropsychiatr Genet. 2005;134b(1):60–66. doi: 10.1002/ajmg.b.30140. [DOI] [PubMed] [Google Scholar]
- 16.Grayson DR, Jia X, Chen Y, et al Reelin promoter hypermethylation in schizophrenia. Proc Natl Acad Sci U S A. 2005;102(26):9341–9346. doi: 10.1073/pnas.0503736102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang HS, Akbarian S. GAD1 mRNA expression and DNA methylation in prefrontal cortex of subjects with schizophrenia. PLoS One. 2007;2(8):e809. doi: 10.1371/journal.pone.0000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murphy BC, O’Reilly RL, Singh SM. Site-specific cytosine methylation in S-COMT promoter in 31 brain regions with implications for studies involving schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2005;133b(1):37–42. doi: 10.1002/ajmg.b.30134. [DOI] [PubMed] [Google Scholar]
- 19.Abdolmaleky HM, Cheng KH, Faraone SV, et al Hypomethylation of MB-COMT promoter is a major risk factor for schizophrenia and bipolar disorder. Hum Mol Genet. 2006;15(21):3132–3145. doi: 10.1093/hmg/ddl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iwamoto K, Bundo M, Yamada K, et al DNA methylation status of SOX10 correlates with its downregulation and oligodendrocyte dysfunction in schizophrenia. J Neurosci. 2005;25(22):5376–5381. doi: 10.1523/JNEUROSCI.0766-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akbarian S, Kim JJ, Potkin SG, et al Gene expression for glutamic acid decarboxylase is reduced without loss of neurons in prefrontal cortex of schizophrenics. Arch Gen Psychiatry. 1995;52(4):258–266. doi: 10.1001/archpsyc.1995.03950160008002. [DOI] [PubMed] [Google Scholar]
- 22.Akbarian S, Huang HS. Molecular and cellular mechanisms of altered GAD1/GAD67 expression in schizophrenia and related disorders. Brain Res Rev . 2006;52(2):293–304. doi: 10.1016/j.brainresrev.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 23.Costa E, Davis J, Grayson DR, Guidotti A, Pappas GD, Pesold C. Dendritic spine hypoplasticity and downregulation of reelin and GABAergic tone in schizophrenia vulnerability. Neurobiol Dis . 2001;8(5):723–742. doi: 10.1006/nbdi.2001.0436. [DOI] [PubMed] [Google Scholar]
- 24.Montano C, Taub MA, Jaffe A, et al Association of DNA methylation differences with schizophrenia in an epigenome-wide association study. JAMA Psychiatry. 2016;73(5):506–514. doi: 10.1001/jamapsychiatry.2016.0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hannon E, Spiers H, Viana J, et al Methylation QTLs in the developing brain and their enrichment in schizophrenia risk loci. Nat Neurosci. 2016;19(1):48–54. doi: 10.1038/nn.4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akbarian S. Epigenetic mechanisms in schizophrenia. Dialogues Clin Neurosci. 2014;16(3):405–417. doi: 10.31887/DCNS.2014.16.3/sakbarian. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang HS, Matevossian A, Jiang Y, Akbarian S. Chromatin immunoprecipitation in postmortem brain. J Neurosci Meth. 2006;156(1-2):284–292. doi: 10.1016/j.jneumeth.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 28.Huang HS, Matevossian A, Whittle C, et al Prefrontal dysfunction in schizophrenia involves mixed-lineage leukemia 1-regulated histone methylation at GABAergic gene promoters. J Neurosci. 2007;27(42):11254–11262. doi: 10.1523/JNEUROSCI.3272-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang B, Dean B, Thomas EA. Disease- and age-related changes in histone acetylation at gene promoters in psychiatric disorders. Transl Psychiatry. 2011;1:e64. doi: 10.1038/tp.2011.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kundakovic M, Jiang Y, Kavanagh DH, et al Practical guidelines for high-resolution epigenomic profiling of nucleosomal histones in postmortem human brain tissue. Biol Psychiatry. 2017;81(2):162–170. doi: 10.1016/j.biopsych.2016.03.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Girdhar K, Hoffman GE, Jiang Y, et al Cell-specific histone modification maps in the human frontal lobe link schizophrenia risk to the neuronal epigenome. Nat Neurosci. 2018;21(8):1126–1136. doi: 10.1038/s41593-018-0187-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu X, Finucane HK, Gusev A, et al Functional architectures of local and distal regulation of gene expression in multiple human tissues. Am J Hum Genet. 2017;100(4):605–616. doi: 10.1016/j.ajhg.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kundaje A, Meuleman W, Ernst J, et al Integrative analysis of 111 reference human epigenomes. Nature. 2015;518(7539):317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sullivan PF, Geschwind DH. Defining the genetic, genomic, cellular, and diagnostic architectures of psychiatric disorders. Cell. 2019;177(1):162–183. doi: 10.1016/j.cell.2019.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roussos P, Mitchell AC, Voloudakis G, et al A role for noncoding variation in schizophrenia. Cell Rep. 2014;9(4):1417–1429. doi: 10.1016/j.celrep.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gusev A, Lee SH, Trynka G, et al Partitioning heritability of regulatory and cell-type-specific variants across 11 common diseases. Am J Hum Genet. 2014;95(5):535–552. doi: 10.1016/j.ajhg.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fromer M, Roussos P, Sieberts SK, et al Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci. 2016;19(11):1442–1453. doi: 10.1038/nn.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dobbyn A, Huckins LM, Boocock J, et al Landscape of conditional eQTL in dorsolateral prefrontal cortex and co-localization with schizophrenia GWAS. Am J Hum Genet. 2018;102(6):1169–1184. doi: 10.1016/j.ajhg.2018.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10(12):1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fullard JF, Giambartolomei C, Hauberg ME, et al Open chromatin profiling of human postmortem brain infers functional roles for non-coding schizophrenia loci. Hum Mol Genet. 2017;26(10):1942–1951. doi: 10.1093/hmg/ddx103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fullard JF, Hauberg ME, Bendl J, et al An atlas of chromatin accessibility in the adult human brain. Genome Res. 2018;28(8):1243–1252. doi: 10.1101/gr.232488.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bryois J, Garrett ME, Song L, et al Evaluation of chromatin accessibility in prefrontal cortex of individuals with schizophrenia. Nat Commun. 2018;9(1):3121. doi: 10.1038/s41467-018-05379-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He X, Fuller CK, Song Y, et al Sherlock: detecting gene-disease associations by matching patterns of expression QTL and GWAS. Am J Hum Genet. 2013;92(5):667–680. doi: 10.1016/j.ajhg.2013.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu Z, Wu C, Wei P, Pan W. A powerful framework for integrating eQTL and GWAS summary data. Genetics. 2017;207(3):893–902. doi: 10.1534/genetics.117.300270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gusev A, Ko A, Shi H, et al Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet. 2016;48(3):245–252. doi: 10.1038/ng.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gusev A, Mancuso N, Won H, et al Transcriptome-wide association study of schizophrenia and chromatin activity yields mechanistic disease insights. Nat Genet. 2018;50(4):538–548. doi: 10.1038/s41588-018-0092-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mancuso N, Shi H, Goddard P, Kichaev G, Gusev A, Pasaniuc B. Integrating gene expression with summary association statistics to identify genes associated with 30 complex traits. Am J Hum Genet. 2017;100(3):473–487. doi: 10.1016/j.ajhg.2017.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gamazon ER, Wheeler HE, Shah KP, et al A gene-based association method for mapping traits using reference transcriptome data. Nat Genet. 2015;47(9):1091–1098. doi: 10.1038/ng.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huckins LM, Dobbyn A, Ruderfer DM, et al Gene expression imputation across multiple brain regions provides insights into schizophrenia risk. Nat Genet. 2019;51(4):659–674. doi: 10.1038/s41588-019-0364-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barbeira AN, Dickinson SP, Bonazzola R, et al Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat Commun. 2018;9(1):1825. doi: 10.1038/s41467-018-03621-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hauberg ME, Fullard JF, Zhu L, et al Differential activity of transcribed enhancers in the prefrontal cortex of 537 cases with schizophrenia and controls. Mol Psychiatry. 2019;22:1402–1412. doi: 10.1038/s41380-018-0059-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li M, Santpere G, Imamura Kawasawa Y, et al Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science (New York, NY) 2018;362(6420) doi: 10.1126/science.aat7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Skene NG, Bryois J, Bakken TE, et al Genetic identification of brain cell types underlying schizophrenia. Nat Genet. 2018;50(6):825–833. doi: 10.1038/s41588-018-0129-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hotz A, Hellenbroich Y, Sperner J, et al Microdeletion 5q14.3 and anomalies of brain development. Am J Med Genet A. 2013;161A(9):2124–2133. doi: 10.1002/ajmg.a.36020. [DOI] [PubMed] [Google Scholar]
- 55.Mitchell AC, Javidfar B, Pothula V, et al MEF2C transcription factor is associated with the genetic and epigenetic risk architecture of schizophrenia and improves cognition in mice. Mol Psychiatry. 2018;23(1):123–132. doi: 10.1038/mp.2016.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li Z, Chen J, Yu H, et al Genome-wide association analysis identifies 30 new susceptibility loci for schizophrenia. Nat Genet. 2017;49(11):1576–1583. doi: 10.1038/ng.3973. [DOI] [PubMed] [Google Scholar]
- 57.Gandal MJ, Zhang P, Hadjimichael E, et al Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science. 2018;362(6420) doi: 10.1126/science.aat8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Consortium P. Revealing the brain’s molecular architecture. Science. 2018;362(6420):1262–1263. doi: 10.1126/science.362.6420.1262. [DOI] [PubMed] [Google Scholar]
- 59.Whalen S, Truty RM, Pollard KS. Enhancer-promoter interactions are encoded by complex genomic signatures on looping chromatin. Nat Genet . 2016;48(5):488–496. doi: 10.1038/ng.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Won H, de la Torre-Ubieta L, Stein JL, et al Chromosome conformation elucidates regulatory relationships in developing human brain. Nature. 2016;538(7626):523–527. doi: 10.1038/nature19847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Whalen S, Pollard KS. Most chromatin interactions are not in linkage disequilibrium. Genome Res. 2019;29(3):334–343. doi: 10.1101/gr.238022.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rajarajan P, Gil SE, Brennand KJ, Akbarian S. Spatial genome organization and cognition. Nat Rev Neurosci. 2016;17(11):681–691. doi: 10.1038/nrn.2016.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rao SS, Huntley MH, Durand NC, et al A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159(7):1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mifsud B, Tavares-Cadete F, Young AN, et al Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat Genet. 2015;47(6):598–606. doi: 10.1038/ng.3286. [DOI] [PubMed] [Google Scholar]
- 65.Davies JO, Telenius JM, McGowan SJ, et al Multiplexed analysis of chromosome conformation at vastly improved sensitivity. Nat Methods . 2016;13(1):74–80. doi: 10.1038/nmeth.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nagano T, Lubling Y, Stevens TJ, et al Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502(7469):59–64. doi: 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nagano T, Lubling Y, Varnai C, et al Cell-cycle dynamics of chromosomal organization at single-cell resolution. Nature. 2017;547(7661):61–67. doi: 10.1038/nature23001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Flyamer IM, Gassler J, Imakaev M, et al Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature. 2017;544(7648):110–114. doi: 10.1038/nature21711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mumbach MR, Rubin AJ, Flynn RA, et al HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat Methods . 2016;13(11):919–922. doi: 10.1038/nmeth.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Du Z, Zheng H, Huang B, et al Allelic reprogramming of 3D chromatin architecture during early mammalian development. Nature . 2017;547(7662):232–235. doi: 10.1038/nature23263. [DOI] [PubMed] [Google Scholar]
- 71.Diaz N, Kruse K, Erdmann T, et al Chromatin conformation analysis of primary patient tissue using a low input Hi-C method. Nat Commun. 2018;9(1):4938. doi: 10.1038/s41467-018-06961-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lu L, Liu X, Peng J, Li Y, Jin F. Easy Hi-C: A simple efficient protocol for 3D genome mapping in small cell populations. bioRxiv. 2018:245688. [Google Scholar]
- 73.Lieberman-Aiden E, van Berkum NL, Williams L, et al Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326(5950):289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rowley MJ, Nichols MH, Lyu X, et al Evolutionarily conserved principles predict 3D chromatin organization. Mol Cell. 2017;67(5):837–852. doi: 10.1016/j.molcel.2017.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rao SSP, Huang SC, Glenn St Hilaire B, et al Cohesin loss eliminates all loop domains. Cell. 2017;171(2):305–320. doi: 10.1016/j.cell.2017.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rowley MJ, Corces VG. Organizational principles of 3D genome architecture. Nat Rev Genet. 2018;19(12):789–800. doi: 10.1038/s41576-018-0060-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dixon JR, Selvaraj S, Yue F, et al Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485(7398):376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hnisz D, Weintraub AS, Day DS, et al Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science. 2016;351(6280):1454–1458. doi: 10.1126/science.aad9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nora EP, Lajoie BR, Schulz EG, et al Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485(7398):381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dixon JR, Jung I, Selvaraj S, et al Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518(7539):331–336. doi: 10.1038/nature14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vietri Rudan M, Barrington C, Henderson S, et al Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep. 2015;10(8):1297–1309. doi: 10.1016/j.celrep.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stadhouders R, Vidal E, Serra F, et al Transcription factors orchestrate dynamic interplay between genome topology and gene regulation during cell reprogramming. Nat Genet. 2018;50(2):238–249. doi: 10.1038/s41588-017-0030-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bonev B, Mendelson Cohen N, Szabo Q, et al Multiscale 3D genome rewiring during mouse neural development. Cell. 2017;171(3):557–572. doi: 10.1016/j.cell.2017.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tang Z, Luo OJ, Li X, et al CTCF-mediated human 3D genome architecture reveals chromatin topology for transcription. Cell. 2015;163(7):1611–1627. doi: 10.1016/j.cell.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guo Y, Xu Q, Canzio D, et al CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell. 2015;162(4):900–910. doi: 10.1016/j.cell.2015.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489(7414):109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Battle A, Brown CD, Engelhardt BE, Montgomery SB. Genetic effects on gene expression across human tissues. Nature. 2017;550(7675):204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Giusti-Rodriguez PM, Sullivan PF. Schizophrenia and a high-resolution map of the three-dimensional chromatin interactome of adult and fetal cortex. bioRxiv. 2018:406330. [Google Scholar]
- 89.Mitchell AC, Bharadwaj R, Whittle C, et al The genome in three dimensions: a new frontier in human brain research. Biol Psychiatry. 2014;75(12):961–969. doi: 10.1016/j.biopsych.2013.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rajarajan P, Borrman T, Liao W, et al Neuron-specific signatures in the chromosomal connectome associated with schizophrenia risk. Science. 2018;362(6420) doi: 10.1126/science.aat4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rajarajan P, Borrman T, Liao W, et al Neuron-specific signatures in the chromosomal connectome associated with schizophrenia risk. Science (New York, NY) 2018;362(6420):eaat4311. doi: 10.1126/science.aat4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rhie SK, Schreiner S, Witt H, et al Using 3D epigenomic maps of primary olfactory neuronal cells from living individuals to understand gene regulation. Sci Adv. 2018;4(12):eaav8550. doi: 10.1126/sciadv.aav8550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang D, Liu S, Warrell J, et al Comprehensive functional genomic resource and integrative model for the human brain. Science (New York, NY) 2018;362(6420) doi: 10.1126/science.aat8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang Q, Chen R, Cheng F, et al A Bayesian framework that integrates multi-omics data and gene networks predicts risk genes from schizophrenia GWAS data. Nat Neurosci. 2019;22(5):691–699. doi: 10.1038/s41593-019-0382-7. [DOI] [PMC free article] [PubMed] [Google Scholar]