Abstract
Skeletal muscle, the largest part of the total body mass, influences energy and protein metabolism as well as maintaining homeostasis. Herein, we demonstrate that during murine muscle satellite cell and myoblast differentiation, transthyretin (TTR) can exocytose via exosomes and enter cells as TTR- thyroxine (T4) complex, which consecutively induces the intracellular triiodothyronine (T3) level, followed by T3 secretion out of the cell through the exosomes. The decrease in T3 with the TTR level in 26-week-old mouse muscle, compared to that in 16-week-old muscle, suggests an association of TTR with old muscle. Subsequent studies, including microarray analysis, demonstrated that T3-regulated genes, such as FNDC5 (Fibronectin type III domain containing 5, irisin) and RXRγ (Retinoid X receptor gamma), are influenced by TTR knockdown, implying that thyroid hormones and TTR coordinate with each other with respect to muscle growth and development. These results suggest that, in addition to utilizing T4, skeletal muscle also distributes generated T3 to other tissues and has a vital role in sensing the intracellular T4 level. Furthermore, the results of TTR function with T4 in differentiation will be highly useful in the strategic development of novel therapeutics related to muscle homeostasis and regeneration.
Keywords: muscle satellite cell, transthyretin, thyroid hormone, myogenesis, exosomes, skeletal muscle
1. Introduction
Skeletal muscle is comprised of multinucleated myofibers and has excellent regeneration capability, which deteriorates progressively with age, restraining the voluntary functions of daily life. The regenerative capacity is mostly facilitated by muscle satellite or stem cells (MSCs) that reside between the basal lamina and sarcolemma, a distinct ‘niche’ in the muscle fibers [1,2]. MSCs vigorously regulate myofiber growth, and MSC progression is typically regulated by the expression of myogenic transcription factors (Pax3, Pax7, myoblast determination protein; MYOD, and myogenin; MYOG) [3]. After injury, quiescent Pax7+ MSCs are triggered to undergo sequential activation, proliferation and differentiation involving MYOD, Myf5 and MYOG to generate multinucleated myotubes [4]. MSC differentiation is indispensable in the regeneration of skeletal muscle and is typically regulated by multiple signaling pathways and by the interaction of several extracellular matrix components with MSCs. Fibromodulin was reported to have a robust role in muscle regeneration by enhancing the recruitment of MSCs to injury sites [5].
Thyroid hormones (THs, thyroxin; T4 and triiodothyronine; T3) have vital roles in the development of various tissues, as well as in postnatal life, by modulating gene expressions [6,7]. THs regulate the expression of various proteins crucial for muscle development and contractility [8,9,10]. Indeed, the foremost targets of THs are muscles, as they regulate the expression of several genes at the transcriptional level [11,12]. The effects of TH signaling in the development and function of skeletal muscle are the result of a remarkably complex mechanism [11]. Generally, to retain homeostasis, regeneration capability, and development, binding of T3 to thyroid hormone receptors (TR) is essential [13]. TRs are encoded by two genes (THRA and THRB), and alternate splicing of each gene produces TRα1, TRβ1, and TRβ2 receptor subtypes. TRα is the predominant subtype in cardiac and skeletal muscle [14]. TRα has a key role in regulation of heart rate and basal metabolism [15]. Transcription of MYOD is directly regulated by T3 [16]. Therefore, TH signaling can control several events during myogenesis via direct and/or indirect regulation of myogenic gene expression.
Retinoids (synthetic vitamin A derivatives) can influence development and metabolism through nuclear hormone receptors (retinoic acid receptor and retinoid X receptor, RXR). RXR forms heterodimers with retinoic acid, TH, and vitamin D receptors, enhancing transcriptional function on their respective response elements [17]. Three different RXR isoforms (RXRα, β and γ) have been characterized. RXRγ is the dominant isoform in adult heart and skeletal muscle [18].
Exosomes are small (40–100 nm) membrane vesicles of endocytic origin that are released from most cell types into the extracellular environment [19]. Exosomes were first defined in 1983, and interest in these vesicles increased markedly after finding that they contain mRNA and microRNA [20]. Exosomes have been shown to facilitate cellular communication by transporting proteins, cytokines, and nucleic acids and to sustain the normal physiological function of cells [21].
Transthyretin (TTR) is a 55-kDa homotetrameric transporter protein for T4 and retinol-binding protein in the blood [22,23]. The liver is the main contributing organ for TTR synthesis in plasma. TTR null (TTR−/−) mice exhibit a delayed suckling-to-weaning transition, delayed growth, reduced muscle mass, and stunted longitudinal bone growth [24]. Among the transporters existing in blood, thyroxine-binding globulin (TBG) has the highest affinity for T4 and T3 (1.0 × 1010 and 4.6 × 108 M−1, respectively), followed by TTR (7.0 × 107 and 1.4 × 107 M−1) and albumin (7.0 × 105 and 1.0 × 105 M−1) [25]. The binding efficacy of TH distributor proteins determines the transportation times for distribution of THs to tissues, thus, TTR (with transitional affinity), more than TBG, is responsible for instant delivery of THs to tissues [6,25].
Though it is known that human placenta, trophoblasts, JEG-3 and HepG2 cells secrete and internalize TTR [26,27,28], its cellular uptake in skeletal muscle has not been fully described. We have demonstrated that TTR initiates myoblast differentiation by inducing the expression of myogenic genes involved in the early phase of myogenesis and the associated calcium channels [6], and we have elucidated its functional role in maintaining the cellular T4 level. Furthermore, we reported that TTR enhances recruitment of MSCs to the site of injury, thereby regulating muscle regeneration [29]. However, the detailed mechanism of TTR with T4 in MSCs differentiation into muscle cells is unclear. In the current work, we have confirmed TTR secretion and internalization in myoblast cell. We found that TTR uptake and internalization by myoblast cells is increased by T4. By using microarray analysis and other studies, we have elucidated that TTR and TH coordinate with each other to modulate gene expression in muscle growth, development, and homeostasis.
2. Materials and Methods
2.1. Animal Experiments
C57BL/6 male mice were obtained from Daehan Biolink (Dae-Jeon, South Korea) and housed at four per cage in a temperature controlled room under a 12 h light/12 h dark cycle. In the period mice (six weeks) were fed a normal diet containing 4.0% (w/w) total fat (Rodent NIH-31 Open Formula Auto; Zeigler Bros., Inc., Gardners, PA, USA). Gastrocnemius muscle tissues were collected after 10 or 20 weeks. After collection, muscle tissues were fixed and stored at −80 °C until required for RNA and protein extraction or fixed overnight at 4 °C for paraffin-embedded tissue blocks to be used in immunohistochemistry. All experimental were done by following the guidelines issued by the Institutional Animal Care and Use Committees of the Catholic University of Daegu (IACUC-2017-051).
2.2. C2C12 Cell Culture
C2C12 cells (murine myoblast, Korean Cell Line Bank, Seoul, Korea) were cultured in DMEM (HyClone Laboratories, Logan, UT, USA) supplemented with 10% FBS (fetal bovine serum, HyClone Laboratories) and 1% P/S (penicillin/streptomycin, Thermo Fisher Scientific, Waltham, MA, USA) in a humidified 5% CO2 incubator at 37 °C. For differentiation, cells were cultured for two or three days in DMEM + 2% FBS + 1% P/S (serum (+) differentiation media) or DMEM + 1% P/S (serum (−) differentiation media). T4 (50 ng/mL, Sigma Aldrich, St. Louis, MO, USA), I-850 (5 ug/mL, Sigma Aldrich), TTR (0.1 ug/mL, Sigma Aldrich) or bovine serum albumin (BSA,1 mg/mL, Sigma Aldrich) was added to the indicated differentiation medium after two or three days.
2.3. Mouse MSCs Culture
Gastrocnemius and cranial thigh muscles were collected from C57BL male mice (six weeks) and minced, digested with 1% pronase (Roche, Mannheim, Germany) for 1 h at 37 °C, and then centrifuged at 1000× g for 3 min followed by passage of the digested tissue phase through a 100 mm syringe filter (Millipore, Darmstadt, Germany). After centrifugation of the filtrate at 1000× g for 5 min, the pellets were suspended in DMEM + 20% FBS + 1% P/S + 5 ng/mL FGF2 (fibroblast growth factor 2, Miltenyi Biotec GmbH, Auburn, CA, USA), seeded on collagen-coated plates (Corning, Brooklyn, NY, USA), and incubated in a humidified 5% CO2 atmosphere at 37 °C. The medium was changed every day. For induction of MSC differentiation into muscle cells, media were switched to DMEM + 2% FBS + 1% P/S or DMEM + 1% P/S followed by incubation for two days. MSC purity was confirmed with Pax7 protein expression (Santa Cruz Biotechnology, Paso Robles, CA, USA) using immunocytochemistry.
2.4. MTT Assay
C2C12 cells were cultured with DMEM + 10% FBS + 1% P/S for two days for analysis of cell viability. The cells were washed with DMEM and then incubated with 0.5 mg/mL MTT reagent (Sigma Aldrich) for 1 h. After dissolving the formazan crystals with DMSO (Sigma Aldrich), absorbance was measured at 540 nm (Tecan Group Ltd., Männedorf, Switzerland).
2.5. Immunoneutralization
TTR protein neutralization was carried out with TTR-specific antibodies (5 µg/mL, Santa Cruz Biotechnology) for two or three days in DMEM + 2% FBS + 1% P/S or DMEM + 1% P/S differentiation media.
2.6. Exosomes Isolation
Cells were cultured with DMEM + 1% P/S differentiation media. The cells were incubated for two or three days and the media were then collected, centrifuged at 2000× g for 30 min, and the upper phase collected for exosomes isolation. Using a total exosomes isolation reagent (Thermo Fisher Scientific, MA, USA), the exosomes from the upper phase were isolated according to the manufacturer’s protocol. In brief, the media were incubated with the total exosomes isolation reagent at 4 °C overnight and centrifuged at 10,000× g for 60 min. After discarding the supernatant, the pellet was dried at room temperature and suspended in PBS.
Mouse plasma (4 mL) was filtered with a 0.8 um syringe filter (Sartorius, Goettingen, Germany), and the exosomes were then isolated according to the manufacturer’s protocol (exoEasy Maxi Kit, Qiagen, Germantown, MD, USA).
2.7. T4 and T3 Concentration Measurement
An ELISA kit (DRG International, Marburg, Germany) was used to measure the concentration of T4 or T3 hormones. In brief, cell lysates or cultured media with T4 or T3 enzyme conjugate reagent were homogenized and added to specific antibody-coated microtiter plates and then incubated for 60 min at room temperature. After discarding the mixtures, the unbound materials were removed by washing the plates. Substrate solution was added followed by incubation for 20 min. Stop solution was then applied to terminate the reaction. Color intensities were then measured at 450 nm by using a spectrophotometer (Tecan Group Ltd., Switzerland).
2.8. Gene Knockdown
When C2C12 cells confluency reached 30%, 1 ng TTR, TR-α, RXRγ, or fibronectin type III domain containing 5 (FNDC5) shRNA vector (Santa Cruz Biotechnology) and scrambled vector (empty vector as negative control, Santa Cruz Biotechnology) were transfected using plasmid transfection reagent and transfection medium according to the manufacturer’s protocol (Santa Cruz Biotechnology). After three days, transfected cells were selected with puromycin (2 ug/mL, shRNA or scrambled vector is a puromycin selection vector, Santa Cruz Biotechnology). Selected cells were grown to 70% confluence before switching to differentiation media. Knockdown efficiencies were determined by analyzing the expressions of control (scrambled vector transfected cell) and knockdown cells. Supplementary Table S1 shows the sequences of the shRNA constructs.
2.9. RNA Isolation, cDNA Synthesis and RealTime RT-PCR
Trizol reagent (Thermo Fisher Scientific) was used following the manufacturer’s instructions to extract total RNA from cells. Two micrograms of RNA in 20 µL of reaction mixture was employed for the synthesis of 1st strand cDNA with random hexamer and reverse transcriptase at 25 °C for 10 min, 37 °C for 120 min, and 85 °C for 5 min. The cDNA product (2 µL) and gene-specific primers (10 pmole, 2 µL) were used for analysis of real-time RT-PCR (40 cycles), which was performed using a 7500 real-time PCR system with power SYBR Green PCR Master Mix (Thermo Fisher Scientific) as the fluorescence source. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the reference gene. Primer information is presented in Supplementary Table S2.
2.10. RT-PCR
Exosomes RNA was synthesized into cDNA and 2 µL cDNA and gene-specific primers (10 pmole, 2 µL) were used for PCR, which was performed using a 2720 Thermal Cycler PCR machine with PCR Master mix (Genetbio, Daejeon, Korea). The PCR conditions were as follow; denaturation 95 °C for 30 s, annealing at 59 °C for 30 s, extension at 72 °C, post-extension 72 °C for 5 min followed by holding (40 cycles). The PCR product was examined by performing electrophoresis on agarose gel.
2.11. Protein Isolation from Culture Media
The cells were cultured with DMEM + 1% P/S differentiation media for two days, centrifuged at 5000× g for 5 min, and the supernatant was then incubated with 1.3% potassium acetate (Sigma Aldrich) for 1 h at 4 °C. The mixture was centrifuged at 1500× g for 10 min, the supernatant was discarded, and the pellet washed with 100% acetone (Merck, Darmstadt, Germany). The isolated proteins were then dried, and Western blot analysis was performed by adding buffer with protease inhibitor cocktail (Thermo Fisher Scientific).
2.12. Western Blot
After washing the cells with PBS, they were lysed with RIPA buffer supplemented with protease inhibitor cocktail (Thermo Fisher Scientific). The Bradford assay was used to estimate the total protein concentration. Proteins (60 µg) were electrophoresed in 10% or 12% SDS-polyacrylamide gel and then transferred to PVDF membrane (EMS–Millipore, Billerica, MA, USA). The blots were then blocked with 3% skim milk or BSA in Tris-buffered saline (TBS)-Tween 20 for 1 h, incubated overnight with protein-specific primary antibodies [TTR (1:400), MYOD (1:500), MYOG (1:500), D2 (iodothyronine deiodinase type 2; 1:500), RXRγ (1:500) (Santa Cruz Biotechnology) or β-actin (1:2000) antibody (Santa Cruz Biotechnology), TR-α (1:500, Thermo Fisher Scientific), MYL2 (myosin light chain 2, 1:1000, Abcam, Cambridge, MA, USA) or FNDC5 (1:500, Bioss Antibodies, Woburn, MA, USA)] in 1% skim milk or BSA in TBS at 4 °C. The blots were then washed and incubated with horse radish peroxidase (HRP)-conjugated secondary antibody (Santa Cruz Biotechnology) for 1 h at room temperature and then developed with Super Signal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific). Supplementary Table S3 shows the molecular weight of protein.
2.13. Fusion Index
After washing with PBS, cells were fixed with methanol and then stained with 0.04% Giemsa G250 (Sigma Aldrich). Images were taken randomly of three different sections per dish. The number of nuclei in myotubes and the total number of nuclei in the cells were counted in each field. Fusion indices were calculated by expressing the number of nuclei in the myotubes as percentages of the total numbers of nuclei.
2.14. TTR Protein Labeling with Fluorescence
TTR protein and BSA were labeled with the Alexa Fluor 594 protein labeling kit (Thermo Fisher Scientific) following the manufacturer’s instructions. Briefly, 100 µL TTR proteins and BSA (0.1 µg/µL) were incubated with 4.7 µL Alexa Fluor 594 succinimidyl ester (12.2 nmole/µL) for 15 min at room temperature, and the conjugated reaction mixture was then purified with resin gel-spin filter. Labeled TTR proteins (0.2 µg) and BSA were added to the cells and detected by fluorescence microscope (Nikon, Tokyo, Japan).
2.15. TTR Overexpression Vector
The region corresponding to the TTR gene open reading frame (ORF) was PCR amplified with TTR ORF primer (Forward: 5′-ATGGCTTCCCTTCGACTCTTCC-3′, Reverse: 5′-GATTCTGGGGGTTGCTGACGA-3′) and ligated into the pcDNA 3.1/CT-GFP-TOPO vector (Invitrogen, Waltham, MA, USA). The ligated sequence was confirmed by sequencing analysis. The construct (2.5 µg) was transfected using 10 µL lipofectamine (1 mg/mL) and Opti MEM medium (Invitrogen) into C2C12 cells following the manufacturer’s directions and positive cells were selected using G418 antibiotics (2 µg/mL, AppliChem GmbH, Darmstadt, Germany).
2.16. Immunocytochemistry
The cells were fixed with 4% formaldehyde (Sigma Aldrich) and permeabilized with 0.2% Triton X 100 (Sigma Aldrich). After blocking with 1% normal goat serum (SeraCare Life Sciences, Milford, MA, USA) for 30 min in a humid environment, cells were incubated with primary antibodies [TTR (1:50), MYOD (1:50), MYOG (1:50), MYL2 (1:50), D2 (1:50), RXRγ (1:50), TRα (1:50), or FNDC5 (1:50)] at 4 °C in a humid environment overnight. Secondary antibody (1: 100; Alexa Fluor 594 goat anti-rabbit or anti-mouse; Thermo Fisher Scientific) was applied for 1 h at room temperature. DAPI was used to stain the cells (Sigma-Aldrich) and imaged using a fluorescence microscope equipped with a digital camera (Nikon, Tokyo, Japan).
2.17. Immunohistochemistry
The sections of paraffin-embedded muscle tissue were deparaffinized and hydrated with xylene (Junsei, Tokyo, Japan) and ethanol (Merck), respectively, and endogenous peroxidase activity was quenched in 0.3% H2O2/methanol. The sections were then either stained with hematoxylin and eosin (Thermo Fisher Scientific) for morphological observation or blocked with 1% normal goat serum (SeraCare Life Sciences), incubated with primary antibodies [TTR (1:50), D2 (1:50), or FNDC5 (1:50)] overnight at 4 °C, and then incubated with horse radish peroxidase–conjugated secondary antibody (1:100). Positive signals were visualized by adding horse radish peroxidase-conjugated streptavidin (Vector, CA, USA). Nuclei of stained sections were stained with hematoxylin and then dehydrated, mounted, and observed by a light microscope (Leica, Wetzlar, Germany).
2.18. Microarray Analysis
Microarray analysis was conducted with the Agilent Technologies mouse GE4X 44K (V2) chip to determine the differentially expressed genes in wild-type (TTRwt) and TTRkd (TTR knockdown) cells as described previously [30]. Briefly, TTRwt and TTRkd cells were grown in serum (+) differentiation media for two days and RNAs were extracted, synthesized into cDNA with fluorescence using a Low RNA Input Linear Amplification kit (Agilent Technologies, CA, USA) according to the manufacturer’s instructions. A total of three hybridizations were performed, and the statistical relevance of gene expression differences was confirmed by SAM (Standard University, Palo Alto, CA, USA). The significance cut-off was a median false discovery rate ≤5% for the SAM analysis.
2.19. DAVID Analysis
DAVID was performed as described previously [5]. In brief, enriched biological themes in up- and down-regulated gene lists (p ≤ 0.05 and 2 fold≤) were categorized by employing the Gene Ontology (GO) terms of cellular component, molecular function, and biological process in DAVID.
2.20. Statistical Analysis
Mean values of normalized expressions were evaluated by Tukey’s Studentized range test to categorize expressional differences of genes, considering p ≤ 0.05 statistically significant. The real-time RT-PCR data was normalized using glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as the internal standard and was analyzed by one-way ANOVA using PROC GLM in SAS 9.0 (SAS Institute, Cary, NC, USA).
3. Results
3.1. TTR Secretion During Myoblast Differentiation
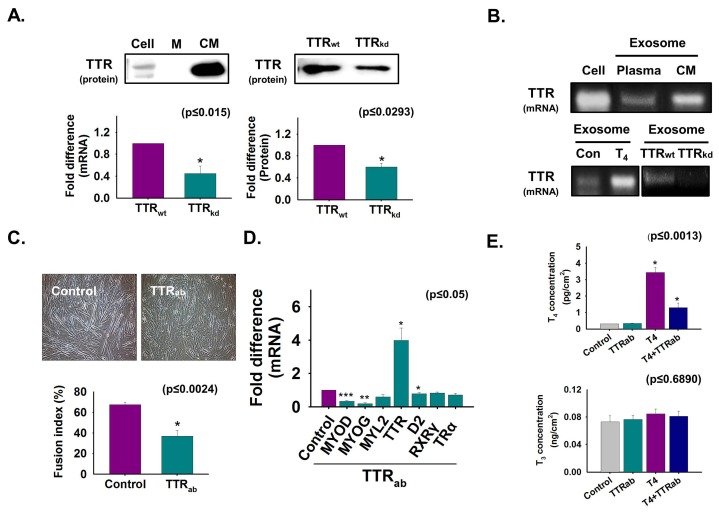
To investigate TTR secretion from cells during C2C12 myoblast differentiation, normal and TTR knockdown cells were cultured in serum-free media for two days, after which the isolated protein level from cultured media was analyzed by Western blotting. The appearance of more TTR protein in cultured media compared to that in cell lysate indicates that TTR is secreted during myoblast differentiation (Figure 1A). Furthermore, TTR mRNA and protein were decreased in TTRkd cells and cultured media, respectively, compared to those in TTRwt cells (Figure 1A). Next, the TTR mRNA level was analyzed in normal cells and exosomes from mouse plasma and media of cultured cells (CM): T4-treated cells, TTRwt, and TTRkd. TTR mRNA was evident in exosomes isolated from culture media and mouse plasma and was increased by T4 treatment but decreased by TTRkd (Figure 1B). TTR immunoneutralization using TTR antibody was performed during differentiation. Myotube formation and the expression of the myogenic genes were decreased by TTR neutralization. However, TTR expression was significantly enhanced in neutralized cells (Figure 1C,D). Interestingly, when the T4 concentration was measured in cells, it was higher in non-neutralized cells than in neutralized cells supplemented with T4 (Figure 1E). Taken together, these results show that TTR secreted from cells transported T4 into the cells during myoblast differentiation.
Figure 1.
The role of secreted TTR from cells during myogenic differentiation. Normal and TTR knockdown cells were cultured with serum-free media for two days (A,B). (A) Proteins were isolated from cells, DMEM (control) and cultured media (CM). TTR protein level was analyzed by Western blot. TTR mRNA level in cells by real-time RT-PCR, and protein level in cell culture media of TTRwt and TTRkd by Western blot. Band intensity was measured by using ImageJ. (B) TTR mRNA levels in normal cell, exosomes isolated from mouse plasma, media of cultured C2C12 cells (CM) with or without T4 treatment, and TTRwt and TTRkd by RT-PCR. Cells were cultured in 2% FBS or serum-free media supplemented with TTR antibody for two (C) or three days (D,E) for immunoneutralization. (C) Myotube formation and fusion index was observed by Giemsa staining. (D) Gene expression was observed by real-time RT-PCR. (E) T4 and T3 concentration in cells was observed by ELISA. TTRwt indicates cells transfected with scrambled vector. Means ± SD (n = 3). * p ≤ 0.05, ** p ≤ 0.001, *** p ≤ 0.0001.
3.2. Enhancement of Myoblast Viability and Differentiation by TTR with T4
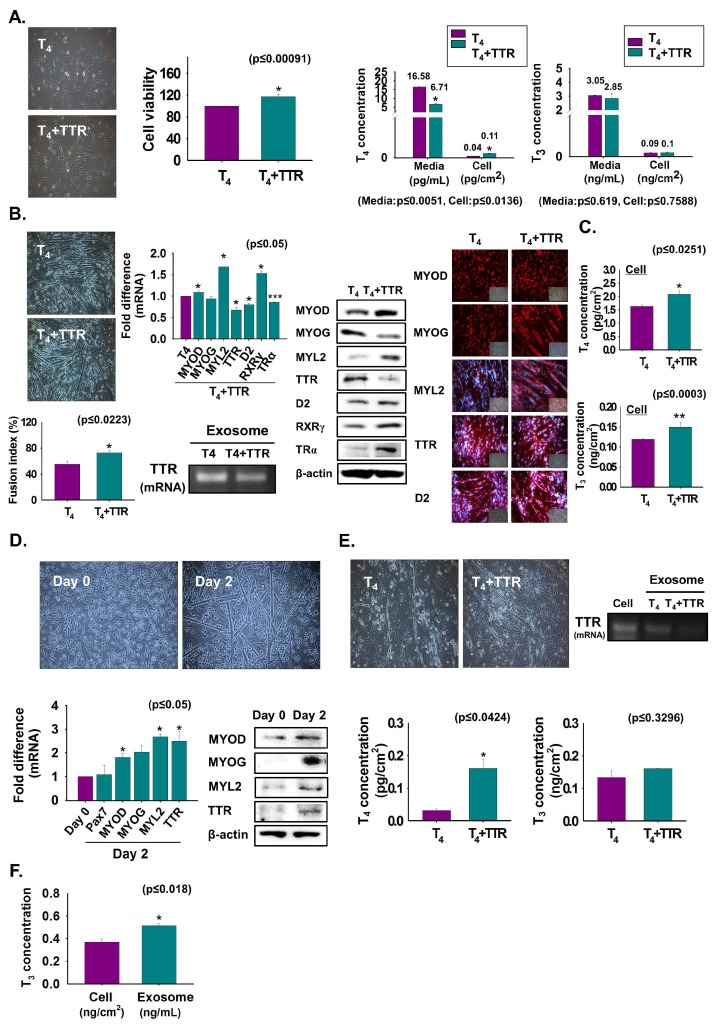
To assess the role of TTR and T4 on myoblast viability and differentiation, C2C12 cells were grown with T4 or T4 + TTR protein for two or three days. Cell viability was increased in T4 + TTR protein treated cells compared to that in only T4 treated cells (Figure 2A). The T4 and T3 concentrations were measured in CM and cells. A lower T4 concentration in media with a consequent higher concentration in cells was observed with T4 + TTR treatment than in those with only T4 treatment. The results indicate that TTR outside the cell enhances the transport of T4 to the cell interior in myoblast viability (Figure 2A). Further, cells were cultured in serum-free media with added T4 or T4 + TTR protein for three days to induce differentiation. The T4 + TTR treatment significantly induced myotube formation with elevated mRNA (MYL2) and protein expression of myogenic factors (MYOD and MYL2), RXRγ, and TRα. However, TTR mRNA and protein expression were decreased by TTR + T4 treatment and their expression in exosomes was also reduced from that of only T4-treated cells (Figure 2B). T4 and T3 concentrations were increased by TTR + T4 treatment (Figure 2C). TTR in mouse MSCs was assessed to determine its expression during differentiation. For this, MSCs were incubated with differentiation media for zero or two days. Expression of TTR and myogenic genes or proteins were increased on day 2 compared to that on day 0 (Figure 2D). Next, MSCs were cultured in serum-free conditions supplemented with T4 or T4 + TTR protein for two days to induce differentiation. Similar to the results with C2C12 cells, MSCs exhibited increased myotube formation with elevated thyroid hormone concentration under T4 + TTR treatment (Figure 2E). Interestingly, decreased TTR mRNA was observed in the exosomes following T4 + TTR treatment (Figure 2E). Furthermore, T3 was present in exosomes isolated from serum-free MSCs culture media supplemented with T4 (Figure 2F). These data showed that TTR protein with T4 not only enhanced myoblast proliferation and myogenic differentiation, but also increased MSC differentiation into muscle cells.
Figure 2.
Myoblast viability and differentiation by treatment with TTR proteins. (A) C2C12 cells were cultured in 10% FBS media supplemented with T4 or T4 + TTR protein for two days. Cell viability was observed by MTT assay. T4 or T3 concentration in cultured media and cells were observed by ELISA. Cells were cultured in serum-free media supplemented with T4 or T4 + TTR protein for three days (B,C). (B) Myotube formation and fusion index by Giemsa staining, mRNA level in cells by real-time RT-PCR, exosomes by RT-PCR, protein expression by Western blot and immunocytochemistry. (C) T4 or T3 concentration in cells was observed by ELISA. (D) When mouse MSCs reached 100% confluency, media were switched to 2% FBS and cultured for zero and two days. MSC differentiation, TTR mRNA level by real-time RT-PCR and protein expression by Western blot. (E) MSCs were cultured in serum-free media supplemented with T4 or T4 + TTR protein for two days. T4 or T3 concentration in cells was observed by ELISA. (F) MSCs were cultured with serum-free media supplemented with T4 for two days and exosomes were isolated from cultured media. T3 concentration in cell and exosomes was observed by ELISA. Means ± SD (n = 3). * p ≤ 0.05, ** p ≤ 0.001, *** p ≤ 0.0001.
3.3. Reduction of T4 Concentration Inside Cells and Myoblast Differentiation by Bovine Albumin Serum (BSA) Treatment
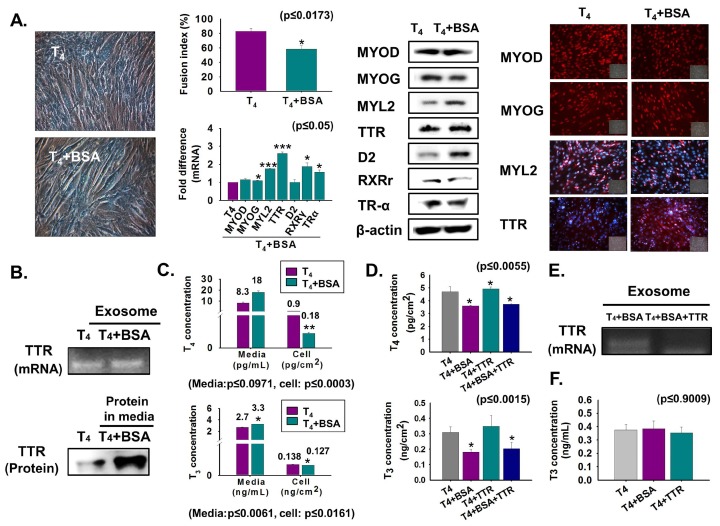
For comparative assessment of T4 transport through TTR to the cell interior, C2C12 cells were cultured in serum-free media supplemented with T4 or T4 + BSA protein for two days. Myotube formation and MYOG expression were decreased in BSA-treated cells, while TTR and D2 expressions were increased at the translational level (Figure 3A). Interestingly, elevated TTR in both exosomes (mRNA) and CM (protein) was also observed in BSA-treated cells (Figure 3B). High T4 and T3 concentrations in T4 + BSA supplemented media with subsequent low levels in both hormone concentrations in the cell, under the same conditions, indicated that BSA reduced the transport of T4 to the cell interior (Figure 3C). Furthermore, elevated T4 concentration was observed in T4 + BSA + TTR supplemented cells relative to that in T4 + BSA treated cells (Figure 3D). Interestingly, decreased TTR mRNA was found in exosomes of T4 + BSA + TTR treated cells (Figure 3E). Additionally, T3 was present in exosomes, and there was no difference in T3 concentration in exosomes supplemented with T4, T4 + BSA, or T4 + TTR (Figure 3F). We observed that BSA reduces myotube formation by decreasing T4 transport.
Figure 3.
Myoblast differentiation following BSA treatment. Cells were cultured in serum-free media supplemented with T4 or T4 + BSA for two days (A–E). (A) Myotube formation and fusion index were observed by Giemsa staining. mRNA level was observed by real-time RT-PCR and protein expressions by Western blot and immunocytochemistry. (B) TTR mRNA in exosomes of cultured media using RT-PCR and protein level in cultured media by Western blot. (C) T4 or T3 concentration in cultured media or cells was observed by ELISA. (D,E) Cells were cultured with serum-free media supplemented with T4, T4 + BSA, T4 + TTR or T4 + BSA + TTR for two days. T4 or T3 concentration in T4 + BSA or T4 + BSA + TTR treated cells. TTR mRNA in exosomes of cultured media (in T4 + BSA or T4 + BSA + TTR treated cells) using RT-PCR. (F) Cells were cultured in serum-free media supplemented with T4 or T4 + BSA or T4 + TTR for two days and exosomes were isolated from each cultured medium. T3 concentration in exosomes. Means ± SD (n = 3). * p ≤ 0.05, ** p ≤ 0.001, *** p ≤ 0.0001.
3.4. TTR Internalization Into Myoblast
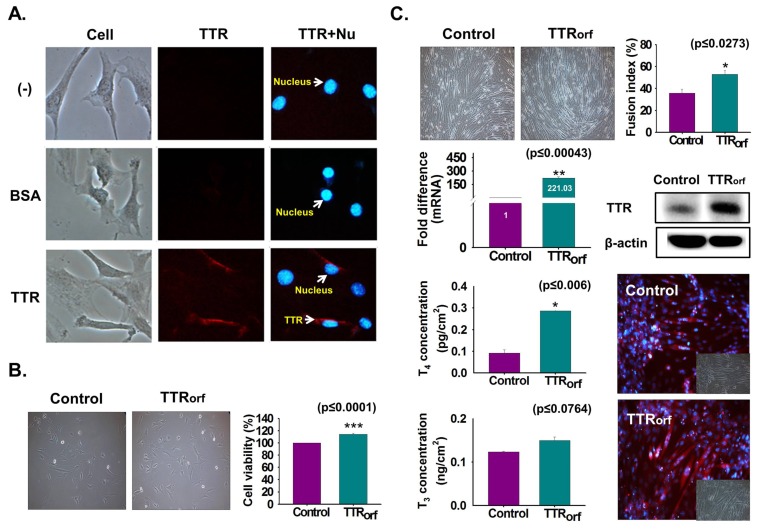
To elucidate TTR internalization to the cell interior, TTR protein or BSA was fluorescently labeled and C2C12 cells were cultured under serum-free conditions supplemented with labeled TTR protein or BSA for one day. Higher fluorescence of labeled TTR protein was evident in the cells treated with labeled TTR than with BSA or in non-treated cells (Figure 4A). TTR overexpression was achieved by transfection with the TTR ORF plasmid and cultured with 10% FBS for two days. Increased cell viability was observed in TTR-overexpressing cells (Figure 4B). Next, TTR-overexpressing cells were cultured with serum-free media for two days. Increased myotube formation with enhanced TTR mRNA/protein expression was observed in TTR-overexpressing cells (Figure 4C). Additionally, elevated concentrations of THs were observed in TTR-overexpressing cells supplemented with T4 (Figure 4C).
Figure 4.
Endocytosis of TTR protein and TTR overexpression effects. (A) TTR protein or BSA were labeled with fluorescence and cells were cultured with serum-free media supplemented with labeled TTR protein or BSA for 1 day. Detection of labeled TTR protein and BSA in cells (Red: TTR, Blue: Nucleus). (B) TTR overexpression was performed by transfecting with TTR ORF plasmid followed by incubation with 10% FBS for two days. Cell viability was analyzed by MTT assay. (C) TTR overexpressing cells were incubated with serum-free media for two days. Myotube formation and fusion index were observed by Giemsa staining, TTR mRNA level by real-time RT-PCR, and protein expression by Western blot and immunocytochemistry. Control or TTR-overexpressing cells were incubated with serum-free media supplemented with T4 for two days. T4 or T3 concentration was measured by ELISA. Means ± SD (n = 3). * p ≤ 0.05, ** p ≤ 0.001, *** p ≤ 0.0001.
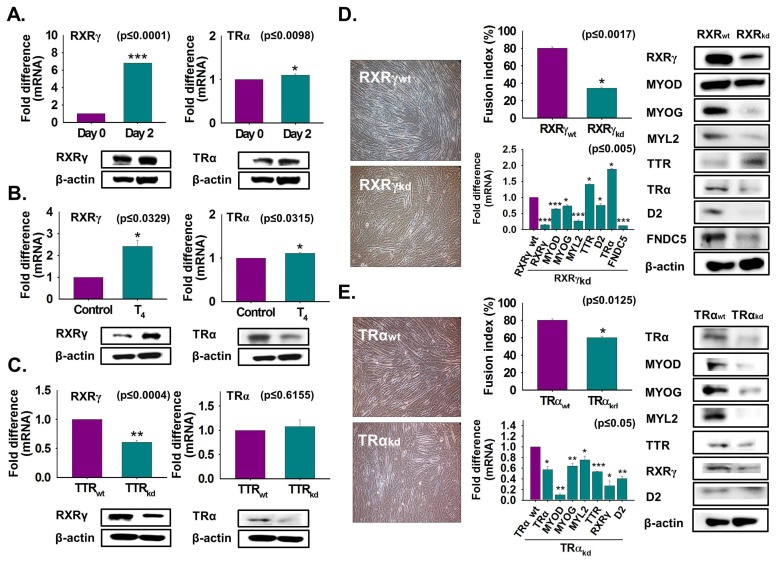
3.5. Regulation of RXRγ and TRα Expression by TTR During Myoblast Differentiation
To determine the role of T4 or TTR on RXRγ and TRα expression, C2C12 cells were grown with or without serum in normal or TTR knockdown cells, and the effects were studied during myoblast differentiation. Increases in mRNA and protein expression of RXRγ and TRα were evident on day 2 compared to the levels on day 0 (Figure 5A). Next, T4 treatment under serum-free conditions stimulated RXRγ expression at both the transcriptional and translational level. However, TRα protein expression was decreased by T4 treatment (Figure 5B). Interestingly, TTR knockdown reduced expression of RXRγ and TRα (Figure 5C). Further, RXRγ and TRα knockdown were performed and followed by culturing with 2% FBS for two days. Myotube formation, myogenic genes and D2 expression were decreased by RXRγ or TRα knockdown, whereas TTR and TRα expressions were increased in RXRγkd cells. Most gene or protein expressions were decreased in TRα knockdown cells (Figure 5D,E). Overall, the above results indicate that expression of RXRγ is controlled by TTR via T4 transportation into the cell during myoblast differentiation.
Figure 5.
RXRγ and TRα expression during myoblast differentiation. (A) Cells were cultured with 2% FBS for two days. RXRγ and TRα expressions using real-time RT-PCR or Western blot. (B) Cells were cultured in serum-free media supplemented with T4 for two days. RXRγ and TRα expression by real-time RT-PCR or Western blot. (C) RXRγ and TRα expression in TTRkd and TTRwt cells using real-time RT-PCR or Western blot. (D) RXRγ knockdown was performed and followed by culture with 2% FBS for two days. Myotube formation and fusion index were observed by Giemsa staining, mRNA expression by real-time RT-PCR, and protein expression by Western blot in RXRγkd and RXRγwt cells. (E) TRα knockdown was performed and followed by culture with 2% FBS for two days. Myotube formation and fusion index were observed by Giemsa staining, mRNA expression by real-time RT-PCR, and protein expression by Western blot in TRαkd and TRαwt cells. TTRwt, RXRγwt, or TRαwt indicate cells transfected with the scrambled vector. Means ± SD (n = 3). * p ≤ 0.05, ** p ≤ 0.001, *** p ≤ 0.0001.
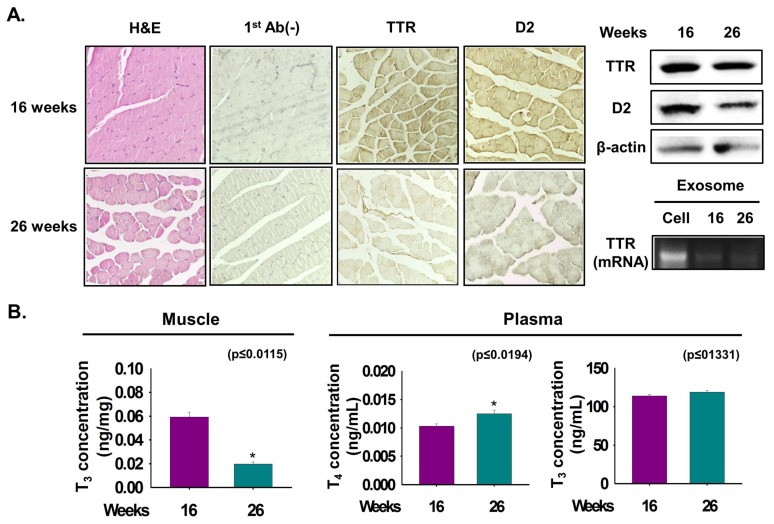
3.6. Relationship between TTR and D2 According to Muscle Age
To determine the effect of muscle age on TTR and D2 expression, mouse muscle at 16- and 26-weeks were collected. Myofiber size (width) and expression of TTR and D2 were decreased in 26-week muscle compared with 16-week muscle (Figure 6A). Interestingly, a decreased TTR level was observed in exosomes isolated from 26-week plasma (Figure 6A). The T3 concentration in 16-week muscle was higher than that in 26-week muscle (Figure 6B). Further, a significant increase in the T4 concentration in the plasma of 26-week mice the was observed, whereas there was no difference in the T3 concentration in the plasma of either age group (Figure 6B). The above findings suggest that expressions of TTR and D2 correlate with the age-dependent differences of muscle.
Figure 6.
TTR expression and T3 concentration in age-dependent differences of muscle.. Expression of TTR and D2 proteins were analyzed in 16- and 26-week mouse muscles. (A) TTR and D2 proteins expression by Immunohistochemistry and Western blot. Exosomes were isolated from 16- and 26-week plasma. TTR mRNA level in cell and exosomes of 16- or 26-week plasma by RT-PCR. (B) T3 or T4 concentration in 16- or 26-week muscles or plasma was observed by ELISA. Means ± SD (n = 3). * p ≤ 0.05, ** p ≤ 0.001, *** p ≤ 0.0001.
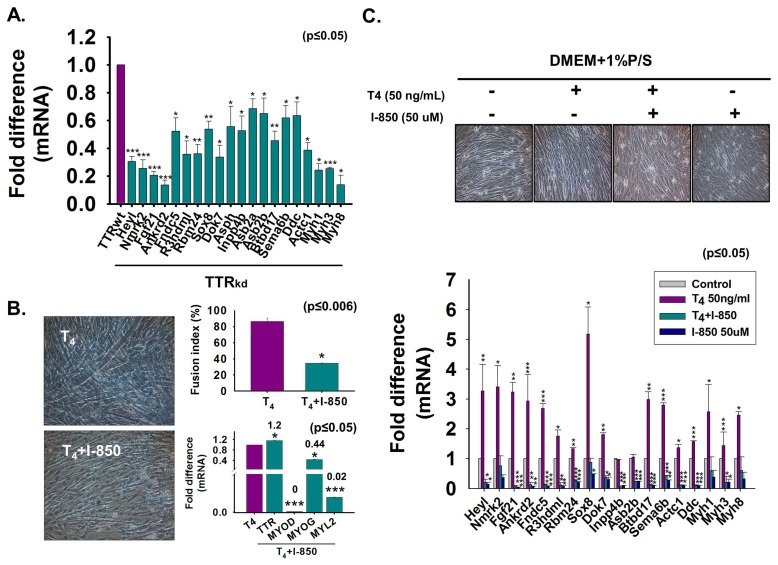
3.7. Microarray Assessment of Gene Expression in TTRkd Cells and Effect of T4 on Gene Expression
To explore TTR function in myoblast differentiation, TTRkd and TTRwt C2C12 cells were cultured with 2% FBS for two days. TTR/MYOG expression and myotube formation were decreased by TTRkd (Supplementary Figure S1A,B). Microarray analysis was performed with TTRwt and TTRkd cells. After applying two-fold cut-offs for down- and up-regulated genes, analysis of the effects of knocking down TTR on myoblasts revealed that, among the genes involved in sarcomere formation, specific genes are actively up- or down-regulated, and some novel genes that are not involved in sarcomere formation functioned at the onset of myogenesis. Among the identified genes, 29 and 7 genes were down- or up-regulated, respectively, by greater than two-fold in TTRkd cells (Table 1A,B; Supplementary Figure S1C,D). Many genes were previously reported to be involved in MSC maintenance (Heyl, Sox8), myogenesis (Fgf21, Ankrd2, Sox8, Asb2), proliferation (Ankrd2), myokine secretion (Fndc5), neuromuscular junction (Dok7), and Ca2+ release of sarcoplasmic reticulum (Asph). Even though some of these genes have identified roles in myogenesis, many novel genes were also affected by TTRkd (R3hdml, Inpp4b, Igf2as, Btbd17, Sema6b and Ddc) (Table 1A; Supplementary Table S4A). However, the upregulated genes were mostly involved in the cell cycle, cell proliferation, and transcription regulation. Interestingly, there was little information indicating that those genes were related to muscle differentiation. Moreover, most of the up-regulated genes were novel genes, but their main functions have been studied in other tissues or organs (Table 1B; Supplementary Table S4B).
Table 1.
Microarray analysis of TTR knockdown.
| A. | |||||||||
| Gene | Set1 | Set2 | Set3 | Set4 | Average | p Value | Description | ||
| Myh1 | 0.16 | 0.31 | 0.13 | 0.06 | 0.16 | 0.0001 | Mus musculus myosin, heavy polypeptide 1, skeletal muscle, adult (Myh1) | ||
| Heyl | 0.25 | 0.19 | 0.15 | 0.1 | 0.17 | 0.0001 | Mus musculus hairy/enhancer-of-split related with YRPW motif-like (Heyl) | ||
| Myo18b | 0.17 | 0.4 | 0.13 | 0.05 | 0.19 | 0.0001 | Mus musculus myosin XVIIIb (Myo18b) | ||
| Myh8 | 0.2 | 0.39 | 0.12 | 0.08 | 0.2 | 0.0001 | Mus musculus myosin, heavy polypeptide 8, skeletal muscle, perinatal (Myh8) | ||
| Nmrk2 | 0.16 | 0.37 | 0.06 | 0.22 | 0.2 | 0.0001 | Mus musculus nicotinamide riboside kinase 2 (Nmrk2) | ||
| Fgf21 | 0.14 | 0.35 | 0.06 | 0.32 | 0.22 | 0.0001 | Mus musculus fibroblast growth factor 21 (Fgf21) | ||
| Ankrd2 | 0.17 | 0.36 | 0.06 | 0.28 | 0.22 | 0.0001 | Mus musculusankyrin repeat domain 2 (stretch responsive muscle) (Ankrd2) | ||
| Myom3 | 0.18 | 0.49 | 0.11 | 0.11 | 0.22 | 0.0001 | Mus musculusmyomesin family, member 3 (Myom3) | ||
| Myh3 | 0.18 | 0.39 | 0.14 | 0.21 | 0.23 | 0.0001 | Mus musculus myosin, heavy polypeptide 3, skeletal muscle, embryonic (Myh3) | ||
| Myh7 | 0.3 | 0.33 | 0.12 | 0.2 | 0.24 | 0.0001 | Mus musculus myosin, heavy polypeptide 7, cardiac muscle, beta (Myh7) | ||
| Myh3 | 0.21 | 0.4 | 0.15 | 0.21 | 0.24 | 0.0001 | Mus musculus myosin, heavy polypeptide 3, skeletal muscle, embryonic (Myh3) | ||
| Fndc5 | 0.44 | 0.26 | 0.18 | 0.1 | 0.25 | 0.0001 | Mus musculus fibronectin type III domain containing 5 (Fndc5) | ||
| R3hdml | 0.35 | 0.4 | 0.09 | 0.15 | 0.25 | 0.0001 | Mus musculus R3H domain containing-like (R3hdml) | ||
| Myh7b | 0.31 | 0.39 | 0.07 | 0.22 | 0.25 | 0.0001 | Mus musculus myosin, heavy chain 7B, cardiac muscle, beta (Myh7b) | ||
| Rbm24 | 0.2 | 0.41 | 0.15 | 0.23 | 0.25 | 0.0001 | Mus musculus RNA binding motif protein 24 (Rbm24) | ||
| Sox8 | 0.45 | 0.21 | 0.13 | 0.22 | 0.25 | 0.0001 | Mus musculus SRY (sex determining region Y)-box 8 (Sox8) | ||
| Dok7 | 0.21 | 0.5 | 0.08 | 0.29 | 0.27 | 0.0002 | Mus musculus docking protein 7 (Dok7) | ||
| Tnnt1 | 0.31 | 0.37 | 0.12 | 0.29 | 0.27 | 0.0001 | Mus musculus troponin T1, skeletal, slow (Tnnt1), transcript variant 1 | ||
| Ttn | 0.17 | 0.48 | 0.24 | 0.2 | 0.27 | 0.0001 | Mus musculus titin (Ttn), transcript variant N2-B | ||
| Asph | 0.28 | 0.46 | 0.18 | 0.16 | 0.27 | 0.0001 | Mus musculus aspartate-beta-hydroxylase (Asph), transcript variant 8 | ||
| Inpp4b | 0.2 | 0.41 | 0.33 | 0.16 | 0.27 | 0.0001 | Mus musculus inositol polyphosphate-4-phosphatase, type II (Inpp4b) | ||
| Igf2os | 0.37 | 0.45 | 0.15 | 0.16 | 0.28 | 0.0001 | Mus musculus insulin-like growth factor 2, opposite strand (Igf2os), antisense RNA | ||
| Myh6 | 0.42 | 0.23 | 0.18 | 0.32 | 0.28 | 0.0001 | Mus musculus myosin, heavy polypeptide 6, cardiac muscle, alpha (Myh6) | ||
| Asb2 | 0.48 | 0.39 | 0.23 | 0.17 | 0.32 | 0.0001 | Mus musculusankyrin repeat and SOCS box-containing 2 (Asb2) | ||
| Mybpc1 | 0.45 | 0.44 | 0.25 | 0.13 | 0.32 | 0.0001 | Mus musculus myosin binding protein C, slow-type (Mybpc1) | ||
| Btbd17 | 0.47 | 0.45 | 0.1 | 0.29 | 0.33 | 0.0002 | Mus musculus BTB (POZ) domain containing 17 (Btbd17) | ||
| Sema6b | 0.46 | 0.39 | 0.09 | 0.38 | 0.33 | 0.0002 | Mus musculussema domain, transmembrane domain (TM), and cytoplasmic domain | ||
| Actc1 | 0.38 | 0.39 | 0.33 | 0.25 | 0.34 | 0.0001 | Mus musculus actin, alpha, cardiac muscle 1 (Actc1) | ||
| Ddc | 0.48 | 0.48 | 0.22 | 0.39 | 0.39 | 0.0001 | Mus musculusdopa decarboxylase (Ddc), transcript variant 1 | ||
| B. | |||||||||
| Gene | Set1 | Set2 | Set3 | Set4 | Average | p Value | Description | ||
| Gm10536 | 4.95 | 7.99 | 11.97 | 13.93 | 9.71 | 0.0049 | Mus musculus predicted gene 10536 (Gm10536), long non-coding RNA | ||
| Iws1 | 3.92 | 4.7 | 2.15 | 10.28 | 5.26 | 0.5130 | Mus musculus IWS1 homolog (S. cerevisiae) (Iws1) | ||
| Dkk2 | 4.02 | 3.54 | 4.95 | 2.98 | 3.87 | 0.0005 | Mus musculusdickkopf homolog 2 (Xenopuslaevis) (Dkk2) | ||
| Cdc45 | 3.44 | 2.22 | 4.92 | 3.16 | 3.43 | 0.0048 | Mus musculus cell division cycle 45 (Cdc45), transcript variant 1 | ||
| Suv420h1 | 3.25 | 2.86 | 2.71 | 3.45 | 3.07 | 0.0001 | Mus musculus suppressor of variegation 4–20 homolog 1 (Drosophila) (Suv420h1) | ||
| Cdc42bpa | 2.28 | 2.11 | 3.11 | 4.32 | 2.95 | 0.0082 | Mus musculus CDC42 binding protein kinase alpha (Cdc42bpa) | ||
| Zfp318 | 2.02 | 2.89 | 2.23 | 4.18 | 2.83 | 0.0094 | Mus musculus zinc finger protein 318 (Zfp318), transcript variant 2 | ||
| C. | |||||||||
| Term | Count | % | p Value | ||||||
| Transcription regulation | 3 | 42.9 | 7.5 × 10−2 | ||||||
| Nucleus | 4 | 57.1 | 9.8 × 10−2 | ||||||
| D. | |||||||||
| Term | Count | % | p Value | ||||||
| Muscle protein | 8 | 28.6 | 1.40 × 10−13 | ||||||
| Thick filament | 6 | 21.4 | 1.00 × 10−12 | ||||||
| Myosin | 7 | 25 | 1.40 × 10−11 | ||||||
| Motor protein | 7 | 25 | 5.90 × 10−9 | ||||||
| Actin-binding | 6 | 21.4 | 8.50 × 10−6 | ||||||
| ATP-binding | 10 | 35.7 | 1.20 × 10−5 | ||||||
| Calmodulin-binding | 5 | 17.9 | 1.80 × 10−5 | ||||||
| Methylation | 6 | 21.4 | 6.60 × 10−5 | ||||||
| Nucleotide-binding | 10 | 35.7 | 8.90 × 10−5 | ||||||
| Coiled coil | 11 | 39.3 | 1.40 × 10−3 | ||||||
| Cytoplasm | 10 | 35.7 | 4.80 × 10−2 | ||||||
| Isopeptide bond | 4 | 14.3 | 9.40 × 10−2 | ||||||
TTRwt or TTRkd were cultured with 2% FBS for two days and microarray analysis was performed on TTRwt or TTRkd. (A and B) List of down- or up-regulated genes in TTRkd (2-fold≤). (C and D) Functional analysis by DAVID (2-fold≤). TTRwt indicates cells transfected with scrambled vector. Means ± SD (n = 3).
Down-regulated genes were analyzed at different myogenic times (0, 2, 4 and 6 days). Interestingly, most gene expressions were increased under myogenic conditions than that at the proliferating stage (Day 0). Similar to MYOG (the myogenic marker gene) the expression of 15 genes increased greatly during myogenic differentiation (Supplementary Figure S2). DAVID analysis was performed using the up- and down-regulated genes. More than half of the up-regulated genes were classified as transcription regulators (Table 1C), especially cell-cycle regulators. Although some of the down-regulated genes were identified as being involved in Ca2+-mediated signal transduction and were reported to regulate transcription, most down-regulated genes were classified as components or regulators of the sarcomere motor unit or the ATPase-related group, which are the main structural components of the sarcomere (Table 1D).
Even though genes were selected based on their high statistical significance among all differentially expressed genes, the genes were also cross-examined by performing real-time RT-PCR with TTRkd and comparing the results to those of TTRwt (Figure 7A). To investigate the effect of T4 on TR expression, cells were grown under serum-free conditions with added T4 and/or TR-specific antagonist 1-850 and examined both for morphological appearance and for changes in mRNA expression levels of certain genes. Myotube formation and mRNA levels of the myogenic marker genes (MYOD, MYOG and MYL2) were decreased by T4 + 1-850 treatment (Figure 7B). In contrast, T4 treatment increased myofibril diameter. The T4 treatment elevated most of the gene mRNA levels, whereas T4 + 1-850 treatment had opposite effects. However, T4 treatment reduced the suppressing effect of 1-850 on mRNA expression of Nmrk2 (40% rescue), Sox8 (40%), Myh1 (20%), and Myh8 (30%) (Figure 7C).
Figure 7.
Expression of down-regulated genes in TTR knock-down cells and effect of T4 treatment on down-regulated genes. (A) TTRwt or TTRkd were cultured with 2% FBS for two days. Down-regulated gene expression was assessed by real-time RT-PCR in TTRwt or TTRkd. (B) Cells were cultured with serum-free media supplemented with T4 or T4 + 1-850 and incubated for two days. Myotube formation and fusion index were observed by Giemsa staining and mRNA expression by real-time RT-PCR. (C) Cells were incubated without or with T4, T4 + 1-850 or 1-850 for two days. Expression of down-regulated genes without or with T4, T4 + 1-850 or 1-850 by real-time RT-PCR. Control indicates non-treated cells. Means ± SD (n = 3). * p ≤ 0.05, ** p ≤ 0.001, *** p ≤ 0.0001.
To determine whether the TTRkd effects were produced by TH and its specific receptor, the TR binding site was scanned in genome portions containing the 5′ flanking region and the first intron of each gene. For precise analysis, two nuclear receptor scanning software programs, NHR-scan and NUBI-scan, were utilized. All binding site candidates were predicted by using the AGGTCA sequence arranged by the DR0, DR4, IR0, IR4, ER4, and ER6 patterns, as was used in the in silico thyroid hormone response elements (TRE) prediction models. Consequently, most of the genes contained more than one TRE at the 5′ flanking region. However, some genes such as Fgf21 did not have a suitable TRE. In addition, Myh1 did not possess a TRE upstream of the first exon (Supplementary Figures S3 and S4). Altogether, these results showed that T4 transported to the cell interior activated TR to induce gene expression and modulated novel and major transcription regulating genes that markedly increased during myogenic differentiation in a TH-dependent manner.
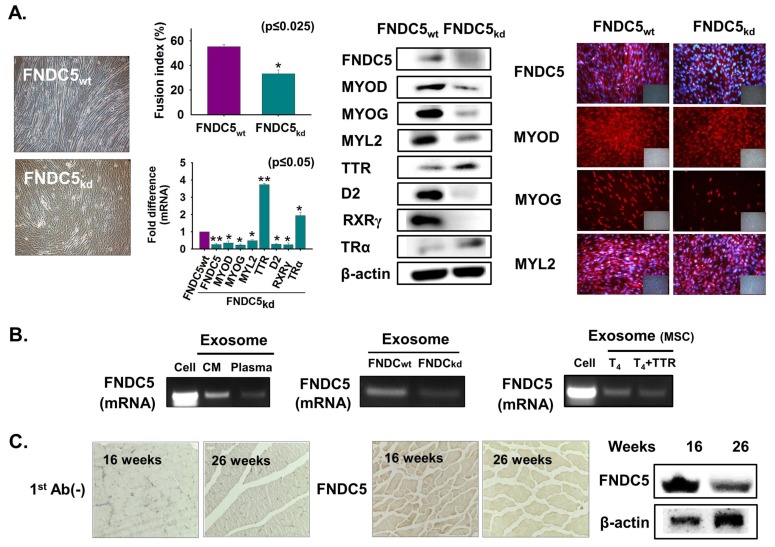
3.8. FNDC5 Expression During Myoblast Differentiation
To confirm the function of the genes that were down-regulated by TTRkd, myokine FNDC5 was selected. FNDC5 knockdown was performed followed by culture with 2% FBS for two days. Myotube formation and myogenic gene expression were decreased in FNDC5kd cells, whereas TTR and TRα expressions were increased at both the transcriptional and translational levels (Figure 8A). Next, cells were grown in serum-free media or supplemented with T4 for two days, and the FNDC5 mRNA level was analyzed in normal cells and exosomes from plasma and media of cultured cells (FNDC5kd and FNDC5wt). FNDC5 mRNA was evident in exosomes from culture media and plasma and decreased in FNDC5kd cells (Figure 8B). Additionally, decreased FNDC5 mRNA was observed in T4 + TTR treatment in MSCs exosomes (Figure 8B). Expression of FNDC5 was decreased in 26-week muscle compared with that in 16-week muscle (Figure 8C). These results show that FNDC5 positively regulates myoblast differentiation.
Figure 8.
FNDC5 expression during myoblast differentiation. (A) FNDC5 knockdown was performed and cells were incubated with 2% FBS for two days. Myotube formation and fusion index were observed by Giemsa staining, mRNA expression using real-time RT-PCR and protein expression were observed by Western blot and immunocytochemistry. (B) Cells were cultured with only serum-free media for two days and exosomes were isolated from cultured media. FNDC5 mRNA level in normal cells, exosomes isolated from plasma, and media of cultured cells (FNDC5wt and FNDC5kd). MSCs were cultured with only serum-free media or supplemented with T4 for two days. FNDC5 mRNA level in exosomes from cell, media of cultured cells with T4 or T4 + TTR. (C) FNDC5 protein expression in 16- or 26-week muscle by immunohistochemistry and Western blot. FNDC5wt indicates cells transfected with scrambled vector. Means ± SD (n = 3). * p ≤ 0.05, ** p ≤ 0.001, *** p ≤ 0.0001.
4. Discussion
Skeletal muscle accounts for nearly half of the body mass and represents the largest protein reservoir in the human body [31]. Although the importance of TH signaling in muscle physiology has been documented for several years, its precise mechanism in skeletal muscle during postnatal myogenesis remains unclear. Initially, we demonstrated the role of TTR in sustaining the cellular T4 level during myoblast proliferation and differentiation [6,29]. In this study, we give the first direct evidence of TTR secretion and uptake in C2C12 mouse myoblast cells. We also identify TTR mRNA in exosomes and its increased expression following T4 treatment, which may act as a mediator in this process. In addition, we studied the role of TTR in T4 transport into C2C12 cells and murine MSCs during the assessment of cell viability and differentiation. The appearance of TTR in cultured serum-free media from myoblasts strongly suggests that TTR synthesized by C2C12 cell is secreted. This suggestion was confirmed by TTR immunoneutralization using TTR antibody, which demonstrated a reduction in myotube formation and mRNA level of some myogenic marker genes (especially, MYOD and MYOG), and T4 uptake into cells, along with an increase in TTR retained in the cells. Further, it is important to emphasize the presence of T3 in exosomes, which indicates that T3 produced in cells is secreted out of the cell through exosomes. These results imply that muscle may not only utilize T4 but also act as a reservoir of T3 in order to distribute it to other tissues or to more distant sites.
Cosmo et al. reported that TH uptake by skeletal muscle can occur independently of monocarboxylate transporter 8 (Mct8). However, they found enhanced TH action, T3 content, and glucose metabolism in Mct8 knockout mice [32]. We speculate that TTR might maintain the TH content in Mct8 knockout mice and, hence, normal muscle metabolism and development. The binding affinity of TTR for T4 is high, hence, it serves as a primary distributor protein in muscle. We showed that TTR with T4 treatment significantly increased cell viability and differentiation compared to that of only T4 treated cells. This was consistent with our previous finding that TTR expression increases myoblast differentiation by increasing T4 transport into the cell [29]. Similar to what we observed in the C2C12 cell line, the T4 + TTR protein treated mouse MSCs also showed increased myotube formation with elevated T4 concentration. Additionally, a progressive increase in TTR expression was observed during differentiation (day 2) in primary MSC cultures. These data confirm that TTR promotes myogenesis by enhancing the transport of T4.
Kassem et al. showed that the availability of TTR in cerebrospinal fluid (CSF) was associated with enhanced T4 uptake into the choroid plexus and brain and this uptake was increased in the presence of TTR [33]. Accordingly, in the present study, low T4 concentration in media with its consequent high concentration in cells supplemented with T4 + TTR indicated that TTR enhanced the transport of T4 to the cell interior during myoblast viability. The enhanced cell uptake may be a simple consequence of the increased T4 level in serum, providing a concentration gradient that promotes TTR secretion and subsequent cell uptake. Furthermore, increased uptake of TH in cells treated with both T4 and TTR probably involves a T4 complex with TTR, as well as passive diffusion of T4, allowing for greater cell uptake than can be accomplished by diffusion only, which is consistent with observations in human ependymoma cells [34]. Although TTR has been reported to be the main component in maintaining high TH levels in CSF and brain [33,35], in this study we observed that TTR also sustained the TH concentration in skeletal muscle and, hence, promoted myogenesis.
TTR is one of three proteins required for T4 transport: TBG is the major transporter and albumin has the lowest affinity, acting as the third T4 binding protein in human plasma [25,36]. Consistent with this theory, we found that BSA reduced T4 transport to the cells, which was also increased with TTR treatment as it has high efficiency for TH. Additionally, BSA treatment decreased myotube formation and myogenic protein expression, while TTR and D2 expressions were increased at the translational level, which might reflect the drop in the T4 level in the cells. TH regulates several genes that are responsible for muscle development and homeostasis. Among those genes, MYOD, MYOG and contractility-determining proteins are transcriptionally regulated by TH and are important for regeneration and myogenesis [37]. MYOD expression regulated by TH is involved in the fast muscle fiber phenotype, with transcriptional stimulation of the myosin-1, myosin-2 and myosin-4 isoforms [38]. TH metabolizing enzyme D2 can activate TH by outer-ring deiodination and can influence local tissue TH levels [39]. Collectively, our findings suggest that TTR acts to maintain the TH level in myoblast cells.
Evidence of high fluorescence-labeled TTR protein levels in cells reveals that TTR was internalized into the myoblast cells. This supports previous results showing endocytosis of fluorescence-labeled TTR in ependymoma cells [34]. In other reports, 125I-TTR and digoxigenin labeled TTR were internalized by an endocytic process in rat yolk sac and β-cells, respectively [40,41]. Furthermore, increased uptake of T4 in TTR-overexpressing cells supplemented with T4 implies that an even distribution of T4 within the cell is not only dependent on the free fraction of T4 in serum but also on the T4 bound to TTR. The presence of T4 or T3 significantly enhanced TTR internalization in JEG-3 cells, with TTR entering the cells as a TTR-T4 complex [27]. In addition, Divino and Schussler [42] reported increased TTR internalization in HepG2 cells with increasing amounts of T4 and suggested that a T4-stimulated conformational alteration in TTR somehow enhanced the uptake of TTR.
Reduced T4 serum concentrations have been reported in old rats [43,44], though their serum T3 level remains more controversial [43]. We show that TTR and D2 expressions with T3 concentration have a correlation with muscle age. The reduced D2 activity is suggestive of impaired T4 conversion in 26-week muscle. Silvestri et al. observed reduced D1 activity in 26-month-old rats relative to that in young (6- and 12-month-old male) rats [44]. Interestingly, decreased TTR expression in 26-week muscle was consistent with the decreased TH transporter Mct8 protein level in liver of 24-month-old rats [44]. Furthermore, higher plasma T3 or T4 concentration in 26-week muscle could be associated with the reduced free T4 concentration in the 16-week muscle, probably due to a higher TBG expression, as described elsewhere [45]. Additionally, decreased T3 concentration in the 26-week muscle at the cellular level was consistent with the findings of Silvestri et al. [44] in which decreased T3 concentration was observed in 24-month-old rats. Nevertheless, T3 generation has been observed in 11-month-old rats relative to that in seven-month-old rats [46], indicating that the mechanisms of T3 production from T4 in old muscle remain poorly understood.
TH is the main endocrine regulator that acts by binding to TRs and imposing a signature type of gene expression [11]. TH primarily functions either via nuclear receptor-mediated stimulation that is T3 dependent or by switching off the gene transcription machinery [13]. In muscle, this signaling pathway is regulated by the THRA1 isoform of TR [47]. The heterodimer complex formed by the TR with RXR- binds to a TRE, leading to activation or suppression of gene transcription [13]. Accordingly, we showed that T4 treatment induced RXRγ expression. However, myotube formation and myogenic factors were decreased in RXRγ and TRα knockdown cells. Interestingly, RXRγ knockout mice are unable to increase their mass in response to high-fat feeding, suggesting a specific effect of RXRγ in skeletal muscle [48]. In muscle, the proteins whose expression are transcriptionally controlled by T3 are SERCA1a [12], SERCA2a [49], uncoupling protein 3 (UCP3) [50], GLUT4 [51], cytosolic malic enzyme (ME1) [52], muscle glycerol-3-phosphate dehydrogenase (mGPDH) [53], and myosin-7 [54]. Furthermore, we found that TTR and D2 expression were decreased in TRαkd cells, which explains the retarded transport of TH into the cell. The selective functions of TRs are controlled by local ligand availability [39,55] or by TH transport to the cell interior via Mct8 or other associated transporters [56]. The TH metabolizing enzymes D2 and D3, as well as transporters Mct8 and Mct10, are expressed in both rodent and human skeletal muscle [57,58].
The TTR-affected genes identified by TTRkd-based microarray analysis included important transcription factors or mediators that have the potential to control several other genes. For example, Rbm24 is reported to regulate MYOG expression [59] and mediate skeletal muscle-specific splicing events [60]. In contrast, Sox8, a negative regulator of myogenesis [61], has increased expression during myogenesis of C2C12 cells. In addition, Sox8 and Heyl genes are marker genes of MSCs [61,62]; however, the Heyl gene showed increased expression during myogenic differentiation. Interestingly, those opposing results were also observed for Nmrk2 [63]. Another research group reported that Ddc is not produced by myotubes [64], but in the present study, it was induced by suitable myogenic differentiation. Altogether, some genes that have been reported to be negatively correlated with myogenesis were markedly increased in expression during myogenic differentiation in this study.
Another interesting observation from the time-course expression study is that several novel genes that show increased expression during myogenesis responded to T4 as they did to TTR. However, Inpp4b and Asb2, genes that contain TREs in proximity to the transcription start site (TSS), did not show any change with T4 treatment. In the case of Inpp4b, TREs in the proximity of the promoter were only downstream of the TSS, and the first intron was approximately 130 kb. This characteristic indicates a rare aspect of the TTRkd-affected genes. Moreover, Fgf21 and Myh1, which do not seem to contain TREs, showed increased expression levels. The various TRE elements have only been predicted by a one-dimensional arrangement, moreover, a proper, precise, and complete nucleotide matrix for this one-dimensional arrangement is not present in public databases. Due to these limitations, many other researchers [65,66] have reported different nucleotide matrices for TREs and different reactivity of each.
Interaction and cooperation between TR and the mammalian insulator CCCTC-binding factor have already been reported [67,68]. An insulator can mediate multi-dimensional chromosomal changes [69]. In addition, based on the results of the TTRkd microarray analysis, the T4 affected sarcomere genes Myh1, Myh3 and Myh8 may be suitable candidates for TR-insulator mediated transcriptional regulation. In the case of the Myh1 gene, no TREs were present in its promoter region.
In contrast, the FNDC5 gene, downregulated by TTRkd, also showed a high expression level during myogenic differentiation and after T4 treatment. The FNDC5 gene encodes the irisin protein, which is considered as a circulating myokine. The most remarkable feature of the FNDC5/irisin protein is that it generates brown fat from white fat [70,71]. Recently, it has been shown that irisin injection stimulated muscle hypertrophy and increased regeneration in injured skeletal muscle [72]. Additionally, enhanced irisin levels have been found during myogenic differentiation and the additional irisin enhances the expression of p-Erk, which has a vital role in the protein synthesis pathway [73]. Thus, knockdown of the FNDC5 gene was undertaken. We showed that interruption of the FNDC5 gene produced a low level of myotube formation. In humans, FNDC5 protein is cleaved to provide detectable irisin levels in circulation. Additionally, increased irisin concentrations occur in response to exercise in humans [74]. Therefore, based on the pro-myogenic role of FNDC5 in the present study, we suggest that FNDC5 may be a potential curative target for the intrusion of muscle dystrophy. Thus, we conclude that one control pathway within TTR myogenesis is mediated by the protein FNDC5.
5. Conclusions
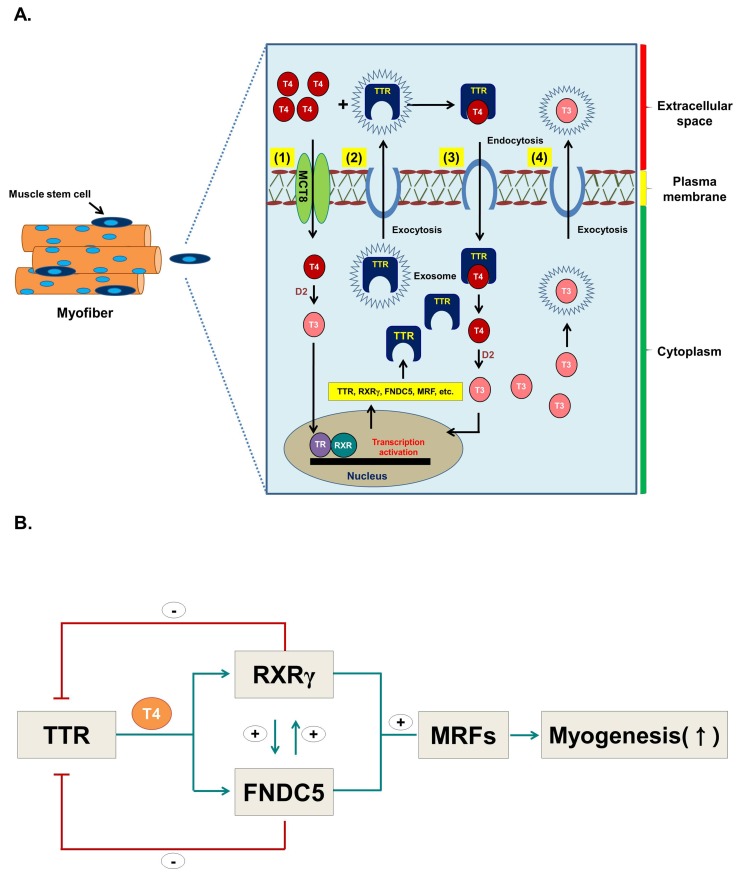
In conclusion, these results suggest that: (1) a portion of the extracellular T4 enters myoblasts or myocytes via MCT via passive diffusion and is converted to T3 by the D2 enzyme which, in turn, induces the expression of several genes including TTR; (2) synthesized TTR exocytoses the cell through exosomes; (3) TTR brings T4 inside the cells as a TTR-T4 complex through an endocytic mechanism; (4) intracellularly synthesized T3 can exocytose via exosomes (Figure 9A); and (5) TTR, through the action of T3 converted from T4, regulates gene expression of TTR intermediates, such as RXRγ and FNDC5 (irisin), which ultimately induces myogenesis (Figure 9B). In this study, we have shown that muscle cells use a much more active mechanism than previously thought to bring T4 into cells. Moreover, intracellularly-generated T3, besides being used in the target muscle cells, also moves out of the cell and affects adjacent cells as well as probably other tissues. Herein, we propose a novel mechanism for the uptake and release of T4 and T3 in myoblasts and for TTR to act as a sensor for intracellular T4 during myogenesis. However, this study has presented a most rudimentary picture of T4 and T3 transport into and out of muscle cells, and further studies will undoubtedly reveal more detailed mechanisms.
Figure 9.
Hypothesis for the role of TTR with T4 during myoblast differentiation. (A) Hypothetical figure depicting role of TTR with T4 during myoblast differentiation. (1) T4 enters cells via Mct8 by passive diffusion and is converted to T3 by D2 enzyme, which in turn triggers the expression of several genes including TTR. (2) Synthesized TTR is exocytosed through exosomes, and (3) subsequently enters the cells as TTR-T4 complex via an endocytic mechanism. (4) T3 produced in the cells can exocytose via exosomes. (B) TTR positively regulates RXRγ and FNDC5 and triggers myogenic regulatory factors, hence promoting myogenesis. RXRγ and FNDC5 negatively regulate TTR while RXRγ and FNDC5 regulate each other.
Abbreviations
| TTR | Transthyretin |
| RXR | Retinoid X receptor |
| MSCs | Muscle satellite cells |
| MYOG | Myogenin |
| T4 | Thyroxin |
| T3 | Triiodothyronine |
| THs | Thyroid hormones |
| TR | Thyroid hormone receptors |
| MYOD | Myoblast determination protein |
| TBG | Thyroxine binding globulin |
| FBS | Fetal bovine serum |
| P/S | Penicillin/Streptomycin |
| D2 | Iodothyronine deiodinase type 2 |
| TRE | Thyroid hormone response elements |
| Mct8 | Monocarboxylate transporter 8 |
| TSS | Transcription start site |
| CSF | Cerebrospinal fluid |
| BSA | Bovine serum albumin |
| FNDC5 | Fibronectin type III domain containing 5 |
| CM | Culture media MYL2 (Myosin light chain 2) |
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4409/8/12/1565/s1, Figure S1: Microarray analysis of TTRkd cells, Figure S2: Time-course study of down-regulated genes during myoblast differentiation, Figure S3: Promoter of down-regulated genes was analyzed to predict TRE binding site, Figure S4: Promoter of down-regulated genes was analyzed to predict TRE binding site, Table S1: shRNA information, Table S2: Primer information, Table S3: Molecular weight of protein, Table S4: Functional analysis of up- or down-regulated genes affected by TTRkd.
Author Contributions
Conceptualization: E.J.L. and I.C.; formal analysis: Y.-W.K. and I.C.; funding acquisition: E.J.L. and I.C.; investigation: E.J.L. and D.C.; methodology: J.H.L., Y.-H.L. and S.-Y.P.; resources: S.J.P. and S.-Y.P.; writing—original draft: E.J.L., S.S. and I.C.; writing—review and editing: E.J.L., S.S., K.A. and M.H.B.
Funding
This research was supported by the National Research Foundation of Korea (NRF) funded by the Korean government (MSIP: grant no. NRF-2018R1A2B6001020) and a grant from the Next-Generation BioGreen 21 Program (project no. PJ01324701), Rural Development Administration, Republic of Korea.
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- 1.Blau H.M., Cosgrove B.D., Ho A.T. The central role of muscle stem cells in regenerative failure with aging. Nat. Med. 2015;21:854–862. doi: 10.1038/nm.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmad K., Lee E.J., Moon J.S., Park S.Y., Choi I. Multifaceted Interweaving between Extracellular Matrix, Insulin Resistance, and Skeletal Muscle. Cells. 2018;8:332. doi: 10.3390/cells7100148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baig M.H., Jan A.T., Rabbani G., Ahmad K., Ashraf J.M., Kim T., Min H.S., Lee Y.H., Cho W.K., Ma J.Y., et al. Methylglyoxal and Advanced Glycation End products: Insight of the regulatory machinery affecting the myogenic program and of its modulation by natural compounds. Sci. Rep. 2017;7:5916. doi: 10.1038/s41598-017-06067-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang K., Zhang Y., Gu L., Lan M., Liu C., Wang M., Su Y., Ge M., Wang T., Yu Y., et al. Islr regulates canonical Wnt signaling-mediated skeletal muscle regeneration by stabilizing Dishevelled-2 and preventing autophagy. Nat. Commun. 2018;9:5129. doi: 10.1038/s41467-018-07638-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee E.J., Jan A.T., Baig M.H., Ashraf J.M., Nahm S.S., Kim Y.W., Park S.Y., Choi I. Fibromodulin: A master regulator of myostatin controlling progression of satellite cells through a myogenic program. FASEB J. 2016;30:2708–2719. doi: 10.1096/fj.201500133R. [DOI] [PubMed] [Google Scholar]
- 6.Lee E.J., Bhat A.R., Kamli M.R., Pokharel S., Chun T., Lee Y.H., Nahm S.S., Nam J.H., Hong S.K., Yang B., et al. Transthyretin is a key regulator of myoblast differentiation. PLoS ONE. 2013;8:e63627. doi: 10.1371/journal.pone.0063627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mishra A., Zhu X.G., Ge K., Cheng S.Y. Adipogenesis is differentially impaired by thyroid hormone receptor mutant isoforms. J. Mol. Endocrinol. 2010;44:247–255. doi: 10.1677/JME-09-0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ambrosio R., De Stefano M.A., Di Girolamo D., Salvatore D. Thyroid hormone signaling and deiodinase actions in muscle stem/progenitor cells. Mol. Cell Endocrinol. 2017;459:79–83. doi: 10.1016/j.mce.2017.06.014. [DOI] [PubMed] [Google Scholar]
- 9.Milanesi A., Lee J.W., Yang A., Liu Y.Y., Sedrakyan S., Cheng S.Y., Perin L., Brent G.A. Thyroid Hormone Receptor Alpha is Essential to Maintain the Satellite Cell Niche During Skeletal Muscle Injury and Sarcopenia of Aging. Thyroid. 2017;27:1316–1322. doi: 10.1089/thy.2017.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soukup T., Smerdu V. Effect of altered innervation and thyroid hormones on myosin heavy chain expression and fiber type transitions: A mini-review. Histochem. Cell Biol. 2015;143:123–130. doi: 10.1007/s00418-014-1276-0. [DOI] [PubMed] [Google Scholar]
- 11.Salvatore D., Simonides W.S., Dentice M., Zavacki A.M., Larsen P.R. Thyroid hormones and skeletal muscle--new insights and potential implications. Nat. Rev. Endocrinol. 2014;10:206–214. doi: 10.1038/nrendo.2013.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simonides W.S., Brent G.A., Thelen M.H., van der Linden C.G., Larsen P.R., van Hardeveld C. Characterization of the promoter of the rat sarcoplasmic endoplasmic reticulum Ca2+-ATPase 1 gene and analysis of thyroid hormone responsiveness. J. Biol. Chem. 1996;271:32048–32056. doi: 10.1074/jbc.271.50.32048. [DOI] [PubMed] [Google Scholar]
- 13.Brent G.A. Mechanisms of thyroid hormone action. J. Clin. Investig. 2012;122:3035–3043. doi: 10.1172/JCI60047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lazar M.A. Thyroid hormone receptors: Multiple forms, multiple possibilities. Endocr. Rev. 1993;14:184–193. doi: 10.1210/edrv-14-2-184. [DOI] [PubMed] [Google Scholar]
- 15.Weiss R.E., Murata Y., Cua K., Hayashi Y., Seo H., Refetoff S. Thyroid hormone action on liver, heart, and energy expenditure in thyroid hormone receptor beta-deficient mice. Endocrinology. 1998;139:4945–4952. doi: 10.1210/endo.139.12.6412. [DOI] [PubMed] [Google Scholar]
- 16.Muscat G.E., Mynett-Johnson L., Dowhan D., Downes M., Griggs R. Activation of myoD gene transcription by 3,5,3’-triiodo-L-thyronine: A direct role for the thyroid hormone and retinoid X receptors. Nucleic Acids Res. 1994;22:583–591. doi: 10.1093/nar/22.4.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leid M., Kastner P., Lyons R., Nakshatri H., Saunders M., Zacharewski T., Chen J.Y., Staub A., Garnier J.A., Mader S., et al. Purification, cloning, and RXR identity of the HeLa cell factor with which RAR or TR heterodimerizes to bind target sequences efficiently. Cell. 1992;68:377–395. doi: 10.1016/0092-8674(92)90478-U. [DOI] [PubMed] [Google Scholar]
- 18.Mangelsdorf D.J., Borgmeyer U., Heyman R.A., Zhou J.Y., Ong E.S., Oro A.E., Kakizuka A., Evans R.M. Characterization of three RXR genes that mediate the action of 9-cis retinoic acid. Genes Dev. 1992;6:329–344. doi: 10.1101/gad.6.3.329. [DOI] [PubMed] [Google Scholar]
- 19.Simpson R.J., Jensen S.S., Lim J.W. Proteomic profiling of exosomes: Current perspectives. Proteomics. 2008;8:4083–4099. doi: 10.1002/pmic.200800109. [DOI] [PubMed] [Google Scholar]
- 20.Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J.J., Lötvall J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 21.Jan A.T., Malik M.A., Rahman S., Yeo H.R., Lee E.J., Abdullah T.S., Choi I. Perspective Insights of Exosomes in Neurodegenerative Diseases: A Critical Appraisal. Front. Aging Neurosci. 2017;9:317. doi: 10.3389/fnagi.2017.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson S.M., Connelly S., Fearns C., Powers E.T., Kelly J.W. The transthyretin amyloidoses: From delineating the molecular mechanism of aggregation linked to pathology to a regulatory-agency-approved drug. J. Mol. Biol. 2012;421:185–203. doi: 10.1016/j.jmb.2011.12.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richardson S.J. Cell and molecular biology of transthyretin and thyroid hormones. Int. Rev. Cytol. 2007;258:137–193. doi: 10.1016/S0074-7696(07)58003-4. [DOI] [PubMed] [Google Scholar]
- 24.Monk J.A., Sims N.A., Dziegielewska K.M., Weiss R.E., Ramsay R.G., Richardson S.J. Delayed development of specific thyroid hormone-regulated events in transthyretin null mice. Am. J. Physiol. Endocrinol. Metab. 2013;304:E23–E31. doi: 10.1152/ajpendo.00216.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alshehri B., D’Souza D.G., Lee J.Y., Petratos S., Richardson S.J. The diversity of mechanisms influenced by transthyretin in neurobiology: Development, disease and endocrine disruption. J. Neuroendocrinol. 2015;27:303–323. doi: 10.1111/jne.12271. [DOI] [PubMed] [Google Scholar]
- 26.Blaner W.S., Bonifacio M.J., Feldman H.D., Piantedosi R., Saraiva M.J. Studies on the synthesis and secretion of transthyretin by the human hepatoma cell line Hep G2. FEBS Lett. 1991;287:193–196. doi: 10.1016/0014-5793(91)80049-9. [DOI] [PubMed] [Google Scholar]
- 27.Landers K.A., McKinnon B.D., Li H., Subramaniam V.N., Mortimer R.H., Richard K. Carrier-mediated thyroid hormone transport into placenta by placental transthyretin. J. Clin. Endocrinol. Metab. 2009;94:2610–2616. doi: 10.1210/jc.2009-0048. [DOI] [PubMed] [Google Scholar]
- 28.McKinnon B., Li H., Richard K., Mortimer R. Synthesis of thyroid hormone binding proteins transthyretin and albumin by human trophoblast. J. Clin. Endocrinol. Metab. 2005;90:6714–6720. doi: 10.1210/jc.2005-0696. [DOI] [PubMed] [Google Scholar]
- 29.Lee E.J., Pokharel S., Jan A.T., Huh S., Galope R., Lim J.H., Lee D.M., Choi S.W., Nahm S.S., Kim Y.W., et al. Transthyretin: A Transporter Protein Essential for Proliferation of Myoblast in the Myogenic Program. Int. J. Mol. Sci. 2017;18:115. doi: 10.3390/ijms18010115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee E.J., Jan A.T., Baig M.H., Ahmad K., Malik A., Rabbani G., Kim T., Lee I.K., Lee Y.H., Park S.Y., et al. Fibromodulin and regulation of the intricate balance between myoblast differentiation to myocytes or adipocyte-like cells. FASEB J. 2018;32:768–781. doi: 10.1096/fj.201700665R. [DOI] [PubMed] [Google Scholar]
- 31.Gonzalez-Freire M., Semba R.D., Ubaida-Mohien C., Fabbri E., Scalzo P., Hojlund K., Dufresne C., Lyashkov A., Ferrucci L. The Human Skeletal Muscle Proteome Project: A reappraisal of the current literature. J. Cachexia Sarcopenia Muscle. 2017;8:5–18. doi: 10.1002/jcsm.12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Di Cosmo C., Liao X.H., Ye H., Ferrara A.M., Weiss R.E., Refetoff S., Dumitrescu A.M. Mct8-deficient mice have increased energy expenditure and reduced fat mass that is abrogated by normalization of serum T3 levels. Endocrinology. 2013;154:4885–4895. doi: 10.1210/en.2013-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kassem N.A., Deane R., Segal M.B., Preston J.E. Role of transthyretin in thyroxine transfer from cerebrospinal fluid to brain and choroid plexus. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2006;291:R1310–R1315. doi: 10.1152/ajpregu.00789.2005. [DOI] [PubMed] [Google Scholar]
- 34.Kuchler-Bopp S., Dietrich J.B., Zaepfel M., Delaunoy J.P. Receptor-mediated endocytosis of transthyretin by ependymoma cells. Brain Res. 2000;870:185–194. doi: 10.1016/S0006-8993(00)02413-6. [DOI] [PubMed] [Google Scholar]
- 35.Chen R.L., Kassem N.A., Preston J.E. Dose-dependent transthyretin inhibition of T4 uptake from cerebrospinal fluid in sheep. Neurosci. Lett. 2006;396:7–11. doi: 10.1016/j.neulet.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 36.Palha J.A. Transthyretin as a thyroid hormone carrier: Function revisited. Clin. Chem. Lab. Med. 2002;40:1292–1300. doi: 10.1515/CCLM.2002.223. [DOI] [PubMed] [Google Scholar]
- 37.Bentzinger C.F., Wang Y.X., Dumont N.A., Rudnicki M.A. Cellular dynamics in the muscle satellite cell niche. EMBO Rep. 2013;14:1062–1072. doi: 10.1038/embor.2013.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allen D.L., Sartorius C.A., Sycuro L.K., Leinwand L.A. Different pathways regulate expression of the skeletal myosin heavy chain genes. J. Biol. Chem. 2001;276:43524–43533. doi: 10.1074/jbc.M108017200. [DOI] [PubMed] [Google Scholar]
- 39.Bianco A.C., Salvatore D., Gereben B., Berry M.J., Larsen P.R. Biochemistry, cellular and molecular biology, and physiological roles of the iodothyronine selenodeiodinases. Endocr. Rev. 2002;23:38–89. doi: 10.1210/edrv.23.1.0455. [DOI] [PubMed] [Google Scholar]
- 40.Dekki N., Refai E., Holmberg R., Kohler M., Jornvall H., Berggren P.O., Juntti-Berggren L. Transthyretin binds to glucose-regulated proteins and is subjected to endocytosis by the pancreatic beta-cell. Cell Mol. Life Sci. 2012;69:1733–1743. doi: 10.1007/s00018-011-0899-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sousa M.M., Norden A.G., Jacobsen C., Willnow T.E., Christensen E.I., Thakker R.V., Verroust P.J., Moestrup S.K., Saraiva M.J. Evidence for the role of megalin in renal uptake of transthyretin. J. Biol. Chem. 2000;275:38176–38181. doi: 10.1074/jbc.M002886200. [DOI] [PubMed] [Google Scholar]
- 42.Divino C.M., Schussler G.C. Transthyretin receptors on human astrocytoma cells. J. Clin. Endocrinol. Metab. 1990;71:1265–1268. doi: 10.1210/jcem-71-5-1265. [DOI] [PubMed] [Google Scholar]
- 43.Mariotti S., Franceschi C., Cossarizza A., Pinchera A. The aging thyroid. Endocr. Rev. 1995;16:686–715. doi: 10.1210/edrv-16-6-686. [DOI] [PubMed] [Google Scholar]
- 44.Silvestri E., Lombardi A., de Lange P., Schiavo L., Lanni A., Goglia F., Visser T.J., Moreno M. Age-related changes in renal and hepatic cellular mechanisms associated with variations in rat serum thyroid hormone levels. Am. J. Physiol. Endocrinol. Metab. 2008;294:E1160–E1168. doi: 10.1152/ajpendo.00044.2008. [DOI] [PubMed] [Google Scholar]
- 45.Savu L., Vranckx R., Rouaze-Romet M., Maya M., Nunez E.A., Treton J., Flink I.L. A senescence up-regulated protein: The rat thyroxine-binding globulin (TBG) Biochim. Biophys. Acta. 1991;1097:19–22. doi: 10.1016/0925-4439(91)90017-4. [DOI] [PubMed] [Google Scholar]
- 46.Jang M., DiStefano J.J., 3rd Some quantitative changes in iodothyronine distribution and metabolism in mild obesity and aging. Endocrinology. 1985;116:457–468. doi: 10.1210/endo-116-1-457. [DOI] [PubMed] [Google Scholar]
- 47.Van Mullem A., van Heerebeek R., Chrysis D., Visser E., Medici M., Andrikoula M., Tsatsoulis A., Peeters R., Visser T.J. Clinical phenotype and mutant TRalpha1. N. Engl. J. Med. 2012;366:1451–1453. doi: 10.1056/NEJMc1113940. [DOI] [PubMed] [Google Scholar]
- 48.Haugen B.R., Jensen D.R., Sharma V., Pulawa L.K., Hays W.R., Krezel W., Chambon P., Eckel R.H. Retinoid X receptor gamma-deficient mice have increased skeletal muscle lipoprotein lipase activity and less weight gain when fed a high-fat diet. Endocrinology. 2004;145:3679–3685. doi: 10.1210/en.2003-1401. [DOI] [PubMed] [Google Scholar]
- 49.Hartong R., Wang N., Kurokawa R., Lazar M.A., Glass C.K., Apriletti J.W., Dillmann W.H. Delineation of three different thyroid hormone-response elements in promoter of rat sarcoplasmic reticulum Ca2+ATPase gene. Demonstration that retinoid X receptor binds 5′ to thyroid hormone receptor in response element 1. J. Biol. Chem. 1994;269:13021–13029. [PubMed] [Google Scholar]
- 50.Solanes G., Pedraza N., Calvo V., Vidal-Puig A., Lowell B.B., Villarroya F. Thyroid hormones directly activate the expression of the human and mouse uncoupling protein-3 genes through a thyroid response element in the proximal promoter region. Biochem. J. 2005;386:505–513. doi: 10.1042/BJ20041073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zorzano A., Palacin M., Guma A. Mechanisms regulating GLUT4 glucose transporter expression and glucose transport in skeletal muscle. Acta Physiol. Scand. 2005;183:43–58. doi: 10.1111/j.1365-201X.2004.01380.x. [DOI] [PubMed] [Google Scholar]
- 52.Desvergne B., Petty K.J., Nikodem V.M. Functional characterization and receptor binding studies of the malic enzyme thyroid hormone response element. J. Biol. Chem. 1991;266:1008–1013. [PubMed] [Google Scholar]
- 53.Dummler K., Muller S., Seitz H.J. Regulation of adenine nucleotide translocase and glycerol 3-phosphate dehydrogenase expression by thyroid hormones in different rat tissues. Biochem. J. 1996;317:913–918. doi: 10.1042/bj3170913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morkin E. Control of cardiac myosin heavy chain gene expression. Microsc. Res. Tech. 2000;50:522–531. doi: 10.1002/1097-0029(20000915)50:6<522::AID-JEMT9>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 55.Gereben B., Zavacki A.M., Ribich S., Kim B.W., Huang S.A., Simonides W.S., Zeold A., Bianco A.C. Cellular and molecular basis of deiodinase-regulated thyroid hormone signaling. Endocr. Rev. 2008;29:898–938. doi: 10.1210/er.2008-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Visser W.E., Friesema E.C., Visser T.J. Minireview: Thyroid hormone transporters: The knowns and the unknowns. Mol. Endocrinol. 2011;25:1–14. doi: 10.1210/me.2010-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Friesema E.C., Jansen J., Jachtenberg J.W., Visser W.E., Kester M.H., Visser T.J. Effective cellular uptake and efflux of thyroid hormone by human monocarboxylate transporter 10. Mol. Endocrinol. 2008;22:1357–1369. doi: 10.1210/me.2007-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marsili A., Ramadan W., Harney J.W., Mulcahey M., Castroneves L.A., Goemann I.M., Wajner S.M., Huang S.A., Zavacki A.M., Maia A.L., et al. Type 2 iodothyronine deiodinase levels are higher in slow-twitch than fast-twitch mouse skeletal muscle and are increased in hypothyroidism. Endocrinology. 2010;151:5952–5960. doi: 10.1210/en.2010-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jin D., Hidaka K., Shirai M., Morisaki T. RNA-binding motif protein 24 regulates myogenin expression and promotes myogenic differentiation. Genes Cells. 2010;15:1158–1167. doi: 10.1111/j.1365-2443.2010.01446.x. [DOI] [PubMed] [Google Scholar]
- 60.Cardinali B., Cappella M., Provenzano C., Garcia-Manteiga J.M., Lazarevic D., Cittaro D., Martelli F., Falcone G. MicroRNA-222 regulates muscle alternative splicing through Rbm24 during differentiation of skeletal muscle cells. Cell Death Dis. 2016;7:e2086. doi: 10.1038/cddis.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schmidt K., Glaser G., Wernig A., Wegner M., Rosorius O. Sox8 is a specific marker for muscle satellite cells and inhibits myogenesis. J. Biol. Chem. 2003;278:29769–29775. doi: 10.1074/jbc.M301539200. [DOI] [PubMed] [Google Scholar]
- 62.Yamaguchi M., Murakami S., Yoneda T., Nakamura M., Zhang L., Uezumi A., Fukuda S., Kokubo H., Tsujikawa K., Fukada S. Evidence of Notch-Hesr-Nrf2 Axis in Muscle Stem Cells, but Absence of Nrf2 Has No Effect on Their Quiescent and Undifferentiated State. PLoS ONE. 2015;10:e0138517. doi: 10.1371/journal.pone.0138517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li J., Mayne R., Wu C. A novel muscle-specific beta 1 integrin binding protein (MIBP) that modulates myogenic differentiation. J. Cell. Biol. 1999;147:1391–1398. doi: 10.1083/jcb.147.7.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smith J.L., Patil P.B., Minteer S.D., Lipsitz J.R., Fisher J.S. Possibility of autocrine beta-adrenergic signaling in C2C12 myotubes. Exp. Biol. Med. 2005;230:845–852. doi: 10.1177/153537020523001109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Harbers M., Wahlstrom G.M., Vennstrom B. Transactivation by the thyroid hormone receptor is dependent on the spacer sequence in hormone response elements containing directly repeated half-sites. Nucleic Acids Res. 1996;24:2252–2259. doi: 10.1093/nar/24.12.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weth O., Weth C., Bartkuhn M., Leers J., Uhle F., Renkawitz R. Modular insulators: Genome wide search for composite CTCF/thyroid hormone receptor binding sites. PLoS ONE. 2010;5:e10119. doi: 10.1371/journal.pone.0010119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ali T., Renkawitz R., Bartkuhn M. Insulators and domains of gene expression. Curr. Opin. Genet. Dev. 2016;37:17–26. doi: 10.1016/j.gde.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 68.Lutz M., Burke L.J., LeFevre P., Myers F.A., Thorne A.W., Crane-Robinson C., Bonifer C., Filippova G.N., Lobanenkov V., Renkawitz R. Thyroid hormone-regulated enhancer blocking: Cooperation of CTCF and thyroid hormone receptor. EMBO J. 2003;22:1579–1587. doi: 10.1093/emboj/cdg147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hou C., Zhao H., Tanimoto K., Dean A. CTCF-dependent enhancer-blocking by alternative chromatin loop formation. Proc. Natl. Acad. Sci. USA. 2008;105:20398–20403. doi: 10.1073/pnas.0808506106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bostrom P., Wu J., Jedrychowski M.P., Korde A., Ye L., Lo J.C., Rasbach K.A., Bostrom E.A., Choi J.H., Long J.Z., et al. A PGC1-alpha-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature. 2012;481:463–468. doi: 10.1038/nature10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roca-Rivada A., Castelao C., Senin L.L., Landrove M.O., Baltar J., Belen Crujeiras A., Seoane L.M., Casanueva F.F., Pardo M. FNDC5/irisin is not only a myokine but also an adipokine. PLoS ONE. 2013;8:e60563. doi: 10.1371/journal.pone.0060563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reza M.M., Subramaniyam N., Sim C.M., Ge X., Sathiakumar D., McFarlane C., Sharma M., Kambadur R. Irisin is a pro-myogenic factor that induces skeletal muscle hypertrophy and rescues denervation-induced atrophy. Nat. Commun. 2017;8:1104. doi: 10.1038/s41467-017-01131-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huh J.Y., Dincer F., Mesfum E., Mantzoros C.S. Irisin stimulates muscle growth-related genes and regulates adipocyte differentiation and metabolism in humans. Int. J. Obes. 2014;38:1538–1544. doi: 10.1038/ijo.2014.42. [DOI] [PubMed] [Google Scholar]
- 74.Jedrychowski M.P., Wrann C.D., Paulo J.A., Gerber K.K., Szpyt J., Robinson M.M., Nair K.S., Gygi S.P., Spiegelman B.M. Detection and Quantitation of Circulating Human Irisin by Tandem Mass Spectrometry. Cell Metab. 2015;22:734–740. doi: 10.1016/j.cmet.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.