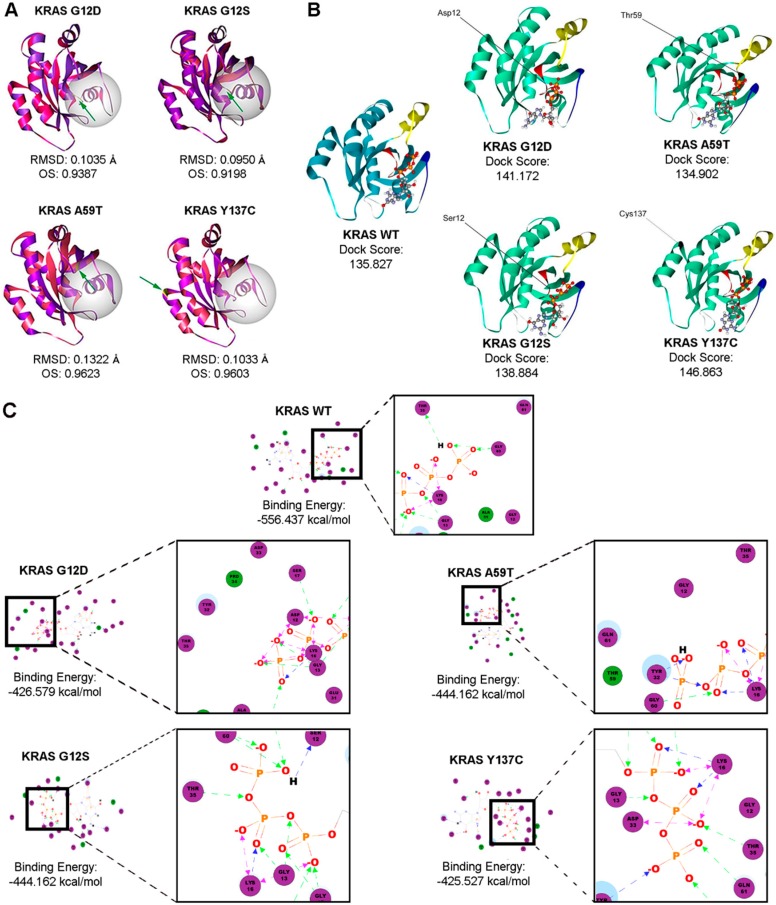
Figure 6.
Bioinformatics-based modeling and docking simulations of the KRAS mutants predict their potential oncogenic impact. (A) KRAS mutants (pink) superimposed with the KRAS wild-type variant (purple) modeled by the Homology Modeler module of the Accelrys Discovery Studio 2.5.5 bioinformatics prediction platform. The position of each mutation is emphasized by the green arrow, while the GTP-binding pocket critical for the GTPase activity of KRAS is highlighted by the white sphere. (B) Docking simulations of wild-type and mutant KRAS with GTP using the LigandFit algorithm. Highlighted in the ribbon structures are the p-loop of KRAS (red), Switch I (yellow), and Switch II (blue), all of which comprise the active site of the KRAS protein. (C) The corresponding 2D-interaction diagrams from the resulting best poses are shown, with the highlighted amino acid interactions inset. One-way green, blue or black arrows between the residue and ligand indicate H-bonding, while two-way pink arrows represent charged interactions. RMSD, root mean square distance; OS, overall similarity; GTP, guanosine triphosphate.