Figure 1.
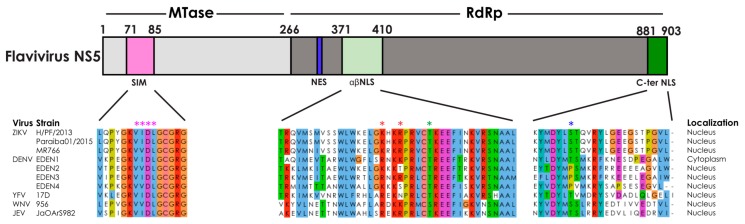
Sequences involved in the regulation of the subcellular localization of flavivirus NS5. (Top) The flavivirus NS5 consists of two functional domains, the methyltransferase (MTase) and the RNA-dependent RNA polymerase (RdRP). The regions involved in its subcellular localization (SIM, NES, αβNLS and C-ter NLS) are also denoted. (Bottom) Amino acid sequence alignment of the denoted regions of NS5 from various flaviviruses showing the sequence conservation using the Clustal X color scheme. The aligned NS5 regions contain the SIM (left), αβNLS (nuclear localization signal) (middle) and C-terminal NLS (right) sequences. The SIM motif is indicated with pink asterisks, two residues mutated in the Zika virus (ZIKV) NS5 NLS mutant are indicated with a red asterisk, CK2 phosphorylated threonine residue is indicated with a green asterisk, and the critical C-terminal NLS residue is indicated with a blue asterisk. The predominant subcellular localization of each NS5 protein is indicated to the right of the alignment. The numbering of amino acid residues is based on H/PF/2013. The virus strains in the alignment and GenBank accession numbers are as follows: H/PF/2013 (KJ776791), Paraiba01/2015 (KX280026), MR766 (LC002520), EDEN1 (EU081230), EDEN2 (EU081177), EDEN3 (EU081190), EDEN4 (GQ398256), YFV (NC 002031.1), WNV (NC_001563.2) and JEV (NC_001437.1).