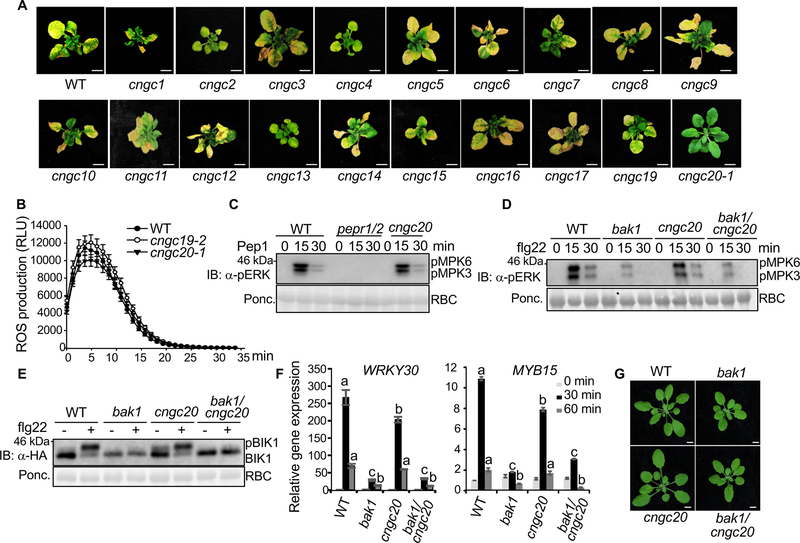
Figure 2. Specific function of CNGC20 in bak1/serk4 cell death.
(A) Unlike cngc20–1, other cngc mutants do not suppress growth defects by RNAi-BAK1/SERK4. Plant phenotypes are shown two weeks after VIGS of BAK1/SERK4. Bar=1 cm.
(B) The cngc19 and cngc20 mutants exhibit similar flg22-induced ROS production as WT plants. Leave discs from four-week-old WT, cngc19–2 and cngc20–1 plants were treated with 100 nM flg22 for 35 min. The data are shown as means ± SE from 24 leaf discs.
(C) The cngc20–3 mutant exhibits similar Pep1-induced MAPK activation as WT. Ten-day-old seedlings were treated without or with 100 nM Pep1 for 15 and 30 min. The MAPK activation was analyzed by immunoblot with α-pERK antibody (top panel), and the protein loading is shown by Ponceau S staining for RBC (bottom panel). The pepr1/2 is the Pep1 receptor PEPR1 and PEPR2 double mutant.
(D) The cngc20–1 mutant does not interfere with the compromised flg22-induced MAPK activation in bak1–4. Ten-day-old seedlings were treated without or with 100 nM flg22 for 15 and 30 min. MAPK activation was analyzed by immunoblotting with α-pERK antibody (top panel), and protein loading is shown by Ponceau S staining for RBC (bottom panel).
(E) The cngc20–1 mutant does not affect the compromised flg22-induced BIK1 phosphorylation in bak1–4. Protoplasts from different plants were transfected with HA-tagged BIK1 and treated with 100 nM flg22 for 10 min. BIK1-HA proteins were detected by immunoblotting using α-HA antibody (top panel), and protein loading is shown by Ponceau S staining for RBC (bottom panel).
(F) The cngc20–1 mutant did not affect the compromised flg22-induced gene expression in bak1–4. Ten-day-old seedlings were treated without or with 100 nM flg22 for 30 or 60 min for qRT-PCR analysis. The data are shown as mean ± SE from three independent repeats. The different letters indicate statistically significant differences from WT within the same time point according to two-way ANOVA followed by Tukey test (p<0.05).
(G) The cngc20–1 mutant does not affect the growth phenotype of bak1–4. Four-week-old soil-grown plants are shown. The cngc20–1/bak1–4 mutant exhibited similar growth phenotypes, including rounder leaves and shorter petioles, as bak1–4 compared to WT plants. Bar=5 mm.
The above experiments were repeated at least three times with similar results.