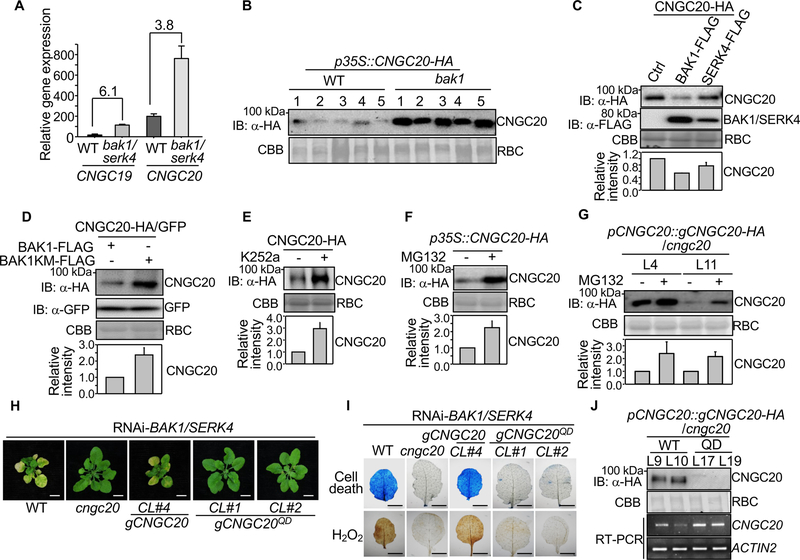
Figure 5. BAK1 destabilizes CNGC20 through phosphorylation.
(A) Transcripts of CNGC19 and CNGC20 are upregulated in bak1–4/serk4–1. Mean gene expression levels of CNGC19 and CNGC20 in bak1–4/serk4–1 and WT were obtained from RNA-Seq data [10]. The data are shown as mean ± SD from two independent repeats. The numbers indicate fold changes of gene expression (bak1–4/serk4–1 vs. WT).
(B) BAK1 is required for CNGC20 destabilization. The CNGC20 cDNA fragment with a C-terminal HA tag under the 35S promoter was introduced into WT or bak1–4. About 20 independent T1 transgenic lines were obtained and five representative lines from each background are shown for immunoblot analysis with α-HA.
(C) CNGC20 proteins are destabilized by co-expression with BAK1 or SERK4. CNGC20-HA was co-expressed with empty vector (Ctrl), BAK1-FLAG or SERK4-FLAG in Arabidopsis protoplasts for 12 hr. Protein expression was analyzed with α-HA or α-FLAG immunoblot.
(D) BAK1, not the BAK1 kinase mutant (BAK1KM), destabilizes CNGC20 proteins. CNGC20-HA or GFP proteins were co-expressed with BAK1-FLAG or BAK1KM-FLAG in Arabidopsis protoplasts for 12 hr. Protein expression was analyzed with α-HA or α-GFP immunoblot.
(E) Pretreatment of kinase inhibitor K252a stabilizes CNGC20-HA in Arabidopsis protoplasts. Protoplasts expressing CNGC20-HA were treated with 0.05% DMSO (−) or 1 μM K252a for 12 hr. Protein expression was analyzed with α-HA immunoblot.
(F) The protein degradation inhibitor MG132 stabilizes CNGC20-HA proteins in N. benthamiana. CNGC20-HA was expressed in N. benthamiana by Agrobacterium-mediated transient assay. 0.1% DMSO (−) or 2 μM MG132 was infiltrated 3 hr before the samples were collected at two days post-inoculation (dpi). Total proteins were analyzed by immunoblot with α-HA.
(G) Stabilization of CNGC20-HA proteins by MG132 in transgenic plants. Ten-day-old pCNGC20::gCNGC20-HA/cngc20–1 seedlings were pretreated with 0.1% DMSO (−) or 2 μM MG132 for 3 hr before total proteins were isolated for immunoblot analysis with α-HA.
Quantifications of CNGC20-HA relative intensity from the immunoblots (IB) are shown on the bottom in (C), (D), (E), (F) and (G) as mean ± SE from three independent repeats. The intensity of CNGC20-HA band normalized to RBC by CBB staining in the first lane in (C) and (D), and the non-treatment in (E), (F) and (G) was set as 1.0.
(H) Complementation of cngc20–1 with the Thr560D/Ser617D/Ser618D/Thr619D quadruple mutant of CNGC20 genomic fragment (gCNGC20QD) under its native promoter does not restore growth defects by RNAi-BAK1/SERK4. CL#1 and CL#2 are two representative lines. Bar=5 mm. CL#4 is the complementation line with WT CNGC20.
(I) The CNGC20QD mutant does not restore cell death and H2O2 production triggered by RNAi-BAK1/SERK4. True leaves of WT, cngc20–1, CNGC20 (CL#4) and CNGC20QD (CL#1 and CL#2) after RNAi-BAK1/SERK4 were stained with trypan blue for cell death (top panel) and DAB for H2O2 accumulation (bottom panel). Bar=2 mm.
(J) CNGC20-HA proteins accumulate more than CNGC20QD-HA proteins in transgenic plants. Plants from two independent homozygous T3 lines of pCNGC20::gCNGC20-HA/cngc20–1 and pCNGC20::gCNGC20QD-HA/cngc20–1 were subjected to immunoblot with α-HA antibody. RT-PCR was performed with CNGC20 specific primers to detect the transcript level of CNGC20 and CNGC20QD. ACTIN2 was used as an internal control.
The above experiments, except A, were repeated three times with similar results.