Figure 2.
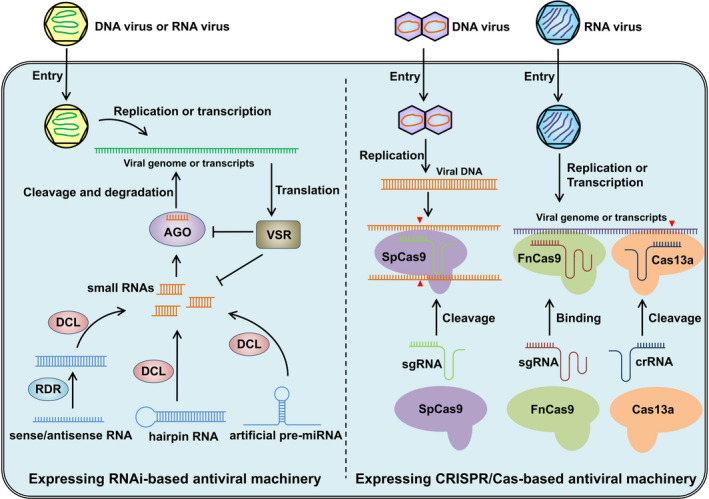
Schematic diagram depicting RNA silencing and CRISPR/Cas strategies to target plant viruses. The diagram on the left shows the mechanism of RNA silencing‐based antiviral engineering. The plant cells are transgenic expressing or exogenous applied of virus‐derived sense/antisense RNA, hairpin RNA, or artificial pre‐miRNA to produce the small RNAs targeting viral genome or transcripts. The small RNAs are loaded into the AGO protein to guide the cleavage of the viral RNA, which induces the degradation of the viral genomic RNA or mRNA. Plant viruses can encode VSR to counter the RNA silencing based resistance by targeting the AGO protein or small RNAs. The diagram on the right shows the mechanism of CRISPR/Cas‐based antiviral engineering. The CRISPR system consists of sgRNA and Cas protein. The transgenic or transit expression of the virus targeting sgRNA and its cognate Cas protein can effectively inhibit the virus infection. Upon DNA virus entry into the plant cell, for example the geminivirus, the viral genome is converted to a doublestranded DNA intermediate, which could be targeted and cleaved by the Cas9 protein from Streptococcus pyogenes (SpCas9). For RNA viruses, Cas9 from Francisella novicida (FnCas9) and Cas13a has been proved to confer virus resistance effectively. Guided by their cognate sgRNA or crRNA, the FnCas9 and Cas13a can bind or cleave the viral genome or transcripts, respectively. Arrowheads in red indicate cleave sites in the viral.
