Figure 1.
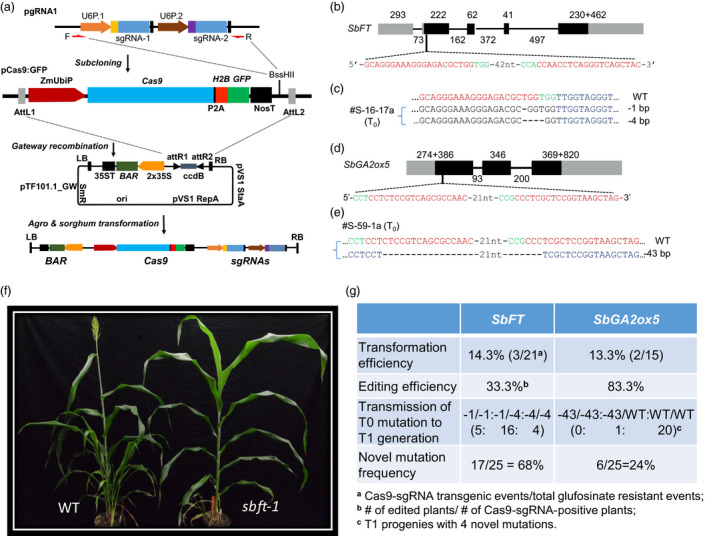
(a) Cloning strategy to construct sorghum CRISPR/Cas9. pgRNA1, a guide RNA cloning vector that contains Btg ZI and BsaI sites for sequential insertions of two double‐stranded oligos for spacer sequences of two sgRNAs. Two different rice U6 promoters are used to drive each of the sgRNAs. The sgRNA cassette was PCR‐amplified with primers F and R; the amplicon is cloned into pCas9:GFP vector at Bss HII site through Gibson assembly. Through Gateway recombination, the Cas9 and sgRNA expression cassettes are mobilized into plant expression vector pTF101.1_GW for Agrobacterium‐mediated sorghum transformation. (b) Gene structure of SbFT on chromosome 10 with sgRNA target sites (in red) and PAM sequences (in green). Forty‐two nucleotides (42nt) indicate sequence not shown between two target sites. Bars in black colour denote coding regions, and bars in grey colour represent untranslated regions. Numbers indicate the nucleotides in exons (above bars) and introns (below lines). (c) Biallelic mutants (1‐bp and 4‐bp deletion) at the first gRNA target site detected in a T0 plant #S‐16‐17a. The nucleotide changes were indicated (dashed lines for deletion and WT for wild type). Dots represent nucleotides not shown. (d) Gene structure of SbGA2ox5 on chromosome 9 with sgRNA target sequences (in red colour) and PAM sequences (in green). Number 21 indicates nucleotides between two target sites not shown. Bars (in solid black) denote coding regions and bars (in grey colour) represent the 5′ and 3′ UTR, respectively. Numbers of nucleotides are denoted as exons (above bars) and introns (below lines). (e) The mono‐allelic mutation (43‐bp deletion) occurring across both gRNA target sites detected in a T0 plant #S‐59‐1a. The nucleotide changes were indicated (dashed lines for deletion and WT for wild type). Dots represent nucleotides not shown. (f) Flowering time was affected by CRISPR‐induced mutation. Morphological characterization of wild type and mutants in the T1 generation. The frameshift mutation created by CRISPR/Cas9 shows the mutant plant (sbft‐1, n = 16, right side) flowered 10 days later on average compared with wild‐type plant (n = 34, left side). (g) Summary of transformation efficiency, CRISPR/Cas9 induced gene‐editing efficiency, transmission of T0 mutation to T1 generation and novel mutations happen in T1 progeny of SbFT and SbGA2ox5.
