Figure 6.
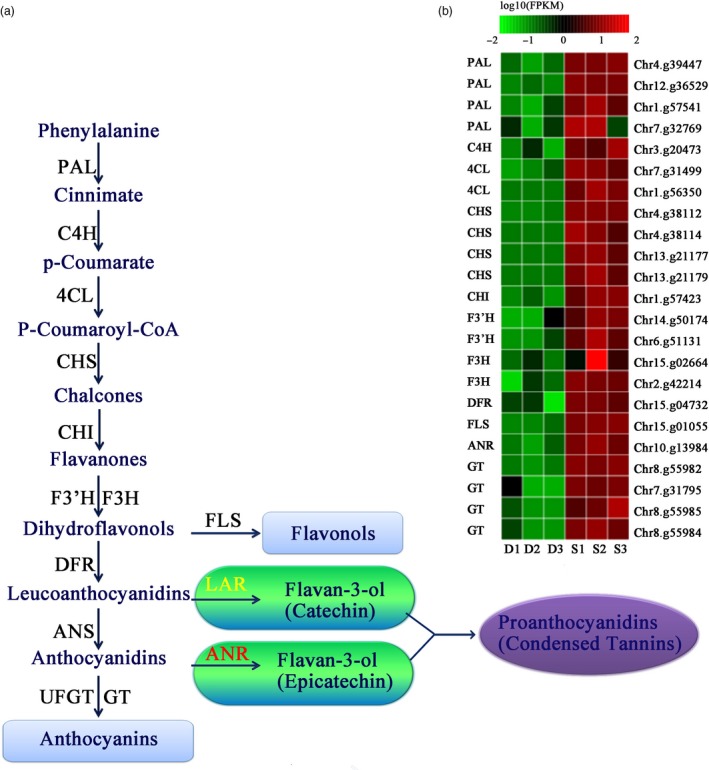
Differential expression of genes involved in proanthocyanidin metabolism in fruits from DSHS and Pbe‐ SD. (A) Proanthocyanidin synthesis pathway and related structural genes in pear fruit. PAL, phenylalanine ammonia lyase; C4H, cinnamate‐4‐hydroxymate; 4CL, 4‐coumarate:coenzyme A ligase; CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone 3‐hydroxylase, F3'H, flavanone 3’‐hydroxylase; DFR, dihydroflavonol 4‐reductase; ANS, anthocyanidin synthase; UFGT, UDP‐glucose flavonoid 3‐O‐glucosyl transferase; FLS, flavonol synthase; LAR, leucoanthocyanidin reductase; ANR, anthocyanidin reductase; GT, glycosyltransferases; and FLS, flavonol synthase. (B) Differentially expressed genes in the proanthocyanidin metabolism pathway. Red colour represents higher than log10 (FPKM) data of genes; green colour represents lower than log10 (FPKM) data of genes; and black colour represents log10 (FPKM) = 0. S1, S2 and S3 indicate the three biological replications of Pbe‐ SD. D1, D2 and D3 indicate the three biological replications of DSHS.
