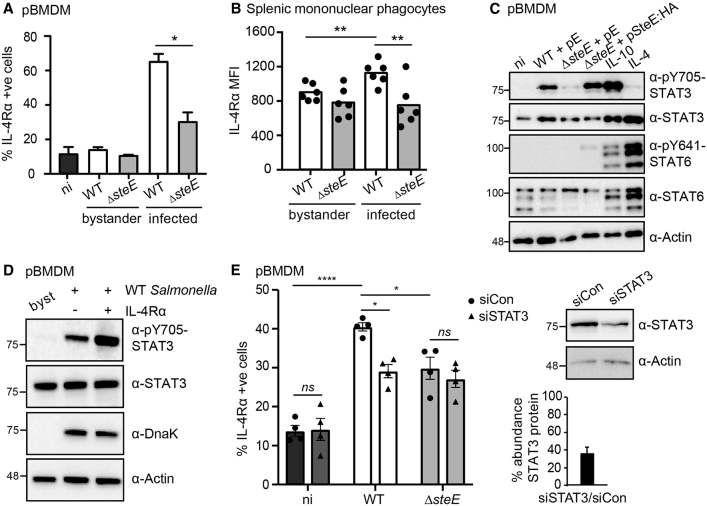
Figure 1.
M2 Macrophage Polarization Is SteE and STAT3 Dependent
(A) Percentage of IL-4Rα+ pBMDMs in naive, non-infected bystander or infected cells 17 h after uptake. Cells were infected with WT or steE mutant Salmonella carrying the fluorescent plasmid pFCcGi (see Figure S1A for gating). Data represent the mean and SEM of four independent experiments (one-way ANOVA with Dunnett’s multiple-comparison test, ∗p < 0.05).
(B) Median fluorescent intensity (MFI) of IL-4Rα signal from CD11b+MHCII+F4/80+ and Ly6G− mononuclear phagocytes isolated from the spleens of C57BL/6 Nramp+/+ mice (see Figure S1B for gating). Mice were infected with WT or steE mutant Salmonella via intraperitoneal inoculation, and spleens were harvested 10 days after infection. Bars represent the geometric median; dots represent individual mice. Significance was calculated with a two-tailed Mann-Whitney test (∗p < 0.05, ∗∗p < 0.01). Data represent three independent experiments with four to six mice analyzed per group per experiment.
(C) Protein immunoblots of whole-cell lysates derived from primary bone-marrow-derived macrophages (pBMDMs) infected with WT, steE mutant, or steE mutant Salmonella carrying a plasmid expressing SteE:HA. pE denotes that the strain carries an empty plasmid. Lysates were harvested 17 h after uptake. Alternatively, pBMDMs were stimulated with 20 mg/mL IL-4 or IL-10 for 17 h, as indicated. Immunoblots are representative of three independent experiments.
(D) pBMDMs were infected with WT Salmonella carrying the fluorescent plasmid pFCcGi for 18 h and then FACS sorted according to whether they were non-infected bystanders (bysts), infected IL4Rα−, or infected IL4Rα+. Only infected cells with a similar bacterial burden were collected (Figure S1C). Sorted cells were then lysed and analyzed by immunoblot with the indicated antibodies. Data are representative of two independent repeats.
(E) Percentage of IL-4Rα+ non-infected or infected pBMDMs 17 h after uptake of WT + pFCcGi or steE mutant + pFCcGi Salmonella after treatment with control (siCon) or STAT3 siRNA for 2 days. Data represent mean and SEM of four independent experiments (two-way ANOVA with Tukey’s multiple-comparison test, ∗p < 0.05, ∗∗p < 0.01; ns, not significant). Inset: immunoblot for STAT3 levels in control or STAT3 siRNA-treated pBMDMs with quantification from three independent repeats shown as mean and SEM.