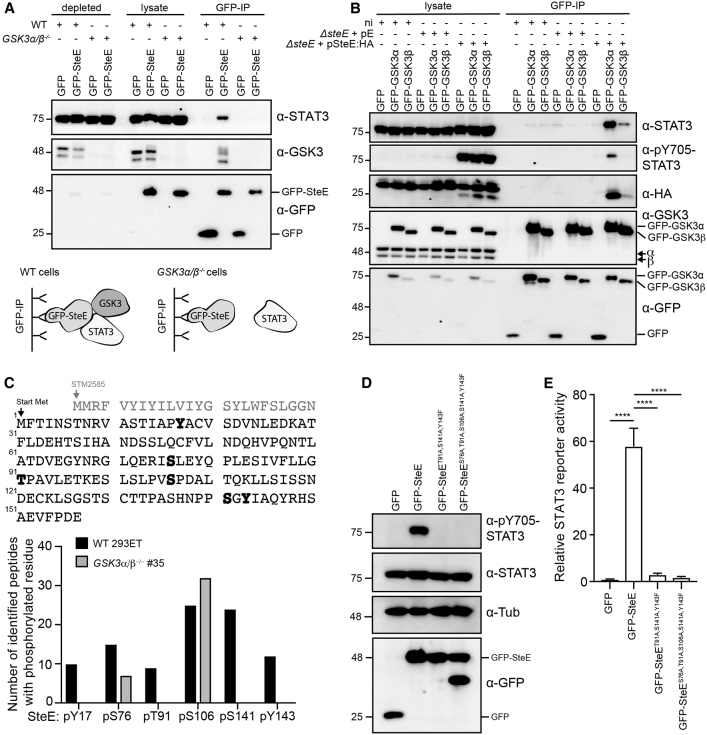
Figure 4.
SteE Enables GSK3 to Form a Complex with STAT3 in which SteE and STAT3 Are Phosphorylated
(A) WT or GSK3α/β−/− 293ET cells expressing GFP or GFP-SteE were lysed, subjected to a GFP immunoprecipitation, and assessed for binding to endogenous STAT3 and GSK3 by immunoblot. Data are representative of three independent repeats.
(B) 293ET cells stably expressing GFP, GFP-GSK3α, or GFP-GSK3β were left non-infected (ni) or infected with steE mutant Salmonella carrying either empty plasmid (pE) or pWSK29-SteE:HA. 17 h after uptake, cell lysates were subject to GFP immunoprecipitation (IP) and lysate, and IP samples were analyzed by immunoblotting for HA, endogenous STAT3, activated STAT3 (pY705), endogenous GSK3, and GFP. Data represent the findings of three independent experiments.
(C) The amino acid sequence and numbering of SteE are shown in black. Amino acids identified as phosphorylated are shown in bold. The number of peptides with each phosphorylated residue, as detected by mass spectrometry, is shown for GFP-SteE expressed in either WT or GSK3α/β−/− 293ET cells. See Figure S5B for protein expression. The data are obtained from three independent repeats.
(D) Whole-cell lysates from 293ET cells transiently expressing GFP, GFP-SteE, or the indicated GFP-tagged SteE mutants were analyzed by immunoblot with antibodies against STAT3, pY705-STAT3, tubulin (Tub), and GFP. Data are representative of three independent experiments.
(E) Luciferase activity in cell lysates from 293ET cells co-transfected with plasmids encoding a STAT3-dependent Firefly luciferase, a constitutively expressed Renilla luciferase, and GFP or the indicated GFP-SteE variant. Data are presented as the fold change in STAT3 reporter activity from GFP-expressing cells and represent the mean and SEM of four independent experiments. Statistical significances were calculated from WT GFP-SteE. ∗∗∗∗p < 0.0001, one-way ANOVA with Dunnett’s post hoc analysis for multiple comparisons.