Figure 5. Linking chaperone recruitment to the structural landscape of domain-specific folding intermediates.
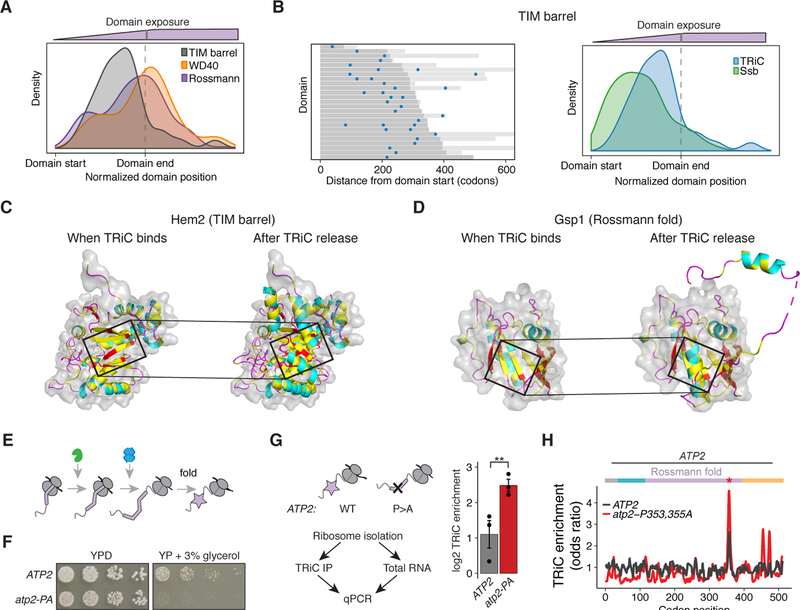
(A) Density distribution of TRiC binding sites in reference to TIM barrels (gray) compared to WD40 domains (orange, P = 5.7e−6) and Rossmann domains (purple, P = 0.002), using the position of peak enrichment for all binding sites.
(B) Heat map of peak enriched position of all TRiC binding sites aligned at the start of TIM barrels (dark gray). Light gray lines indicate the length of the protein beyond the domain end. Density plot shows Ssb sites. n = 34 TRiC sites assigned to 25 domains in 25 ORFs, and 67 Ssb sites assigned to 42 domains in 40 ORFs; P = 0.001, Wilcoxon rank-sum test.
(C – D) Structure of (C) Hem2’s TIM barrel (PDB ID: 1H7N (Erskine et al., 2001)) and (D) Gsp1’s Rossmann fold (PDB ID: 3M1I (Koyama and Matsuura, 2010)) exposed from the ribosome at the time TRiC binds (left) as well as the complete domain after TRiC is released (right). Cartoon colored according to secondary structure, with hydrophobic residues in yellow, and surface representation of just the region exposed before TRiC binds. Box denotes the nascent chain that emerges just before TRiC binds.
(E) Schematic of TRiC binding after Ssb as the end of a domain is translated.
(F) Growth of WT and mutant atp2-P353,355A yeast on YPD or media containing the non-fermentable carbon source of glycerol. Representative image of 5 biological replicates is shown.
(G) TRiC enrichment of ATP2 and atp2-P353,355A. Total and TRiC-bound mRNA was isolated from RNCs followed by qPCR. n = 3 biological replicates with mean ± SEM. P = 0.01, Welch’s t-test.
(H) TRiC enrichment ribosome occupancy profile of ATP2 or atp2-P353,355A.
See also Figure S6.