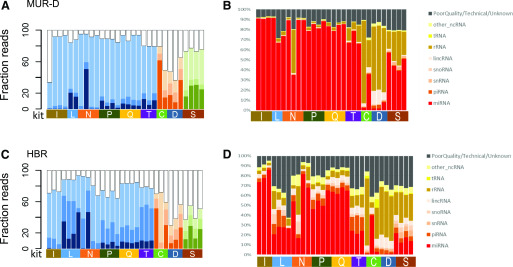
FIGURE 4.
Detection of smRNAs in complex samples. Insert sizes in MUR-D (A) and HBR (B) are shown with inserts <18 nt as white, 18–23 nt as the lightest shade, 24–35 nt as the medium shade, and >35 nt as the darkest shade. Data from the different method types are colored (ligation, blue; polyA tailing, orange; circularization, green). The right panels show the smRNA target distribution for MUR-D (C) and HBR (D). Mapped targets are noted in the key. Unmapped targets are shown in black. C, Takara Bio (Clontech) SMARTer smRNA-Seq Kit; D, Diagenode CATS Small RNA-Seq Kit; I, Illumina TruSeq Small RNA Library Prep Kit; L, Lexogen Small RNA-Seq Library Prep Kit; lincRNA, long intervening noncoding RNA; N, New England Biolabs NEBNext Small RNA Library Prep Set; ncRNA, noncoding RNA; P, PerkinElmer NextFlex Small RNA-Seq Kit v.3; Q, Qiagen QIAseq miRNA Library Kit; S, Somagenics RealSeq-AC miRNA Library Kit; snoRNA, small nucleolar RNA; snRNA, small nuclear RNA; T, Trilink CleanTag Small Library Prep Kit.