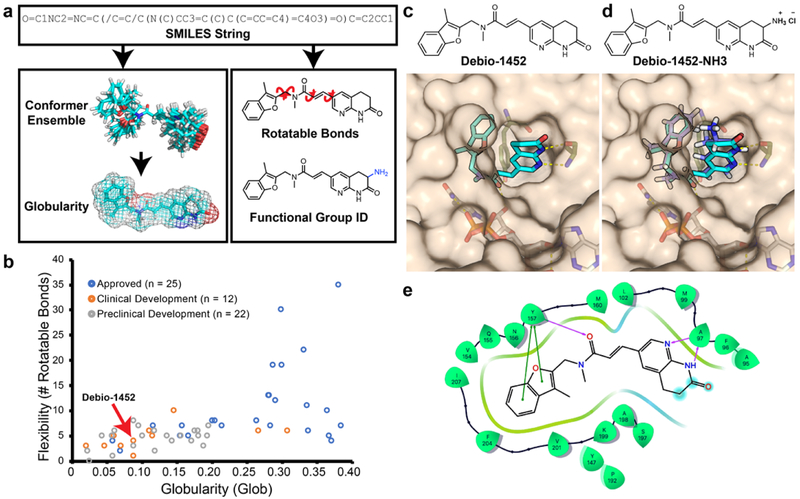
Fig. 1. Overview of eNTRyway molecule processing.
a. Molecules are submitted as SMILES strings, and after an initial estimate of the 3D structure, a conformer space is systematically explored from which an average globularity is calculated. Particular functional groups and number of rotatable bonds are determined directly from 2D structure. b. Physiochemical properties of existing Gram-positive-only antibiotics, Debio-1452 is indicated with red arrow (for specific compounds see Supplementary Table 1). c. Solvent exposure of the naphthyridinone of Debio-1452 (hydrogen atoms are not shown, carbon atoms are shown in cyan, nitrogen atoms are shown in blue, oxygen atoms are shown in red) when bound to S. aureus FabI (PDB: 4FS3) suggests sites for modification. d. Molecular docking of amine-containing derivative, Debio-1452-NH3 (hydrogen atoms are shown in white, carbon atoms are shown in lavender, nitrogen atoms are shown in blue, and oxygen atoms are shown in red). overlaid with the crystal structure of Debio-1452 (hydrogen atoms are not shown, carbon atoms are shown in cyan, nitrogen atoms are shown in blue, oxygen atoms are shown in red). Additional derivative docking scores are shown in Supplementary Table 3, and a non-overlapping pose of Debio-1452-NH3 is shown in Extended Data Fig. 2. e. Ligand interaction diagram of interaction shown in panel c, cyan spheres indicate positions of solvent exposure.