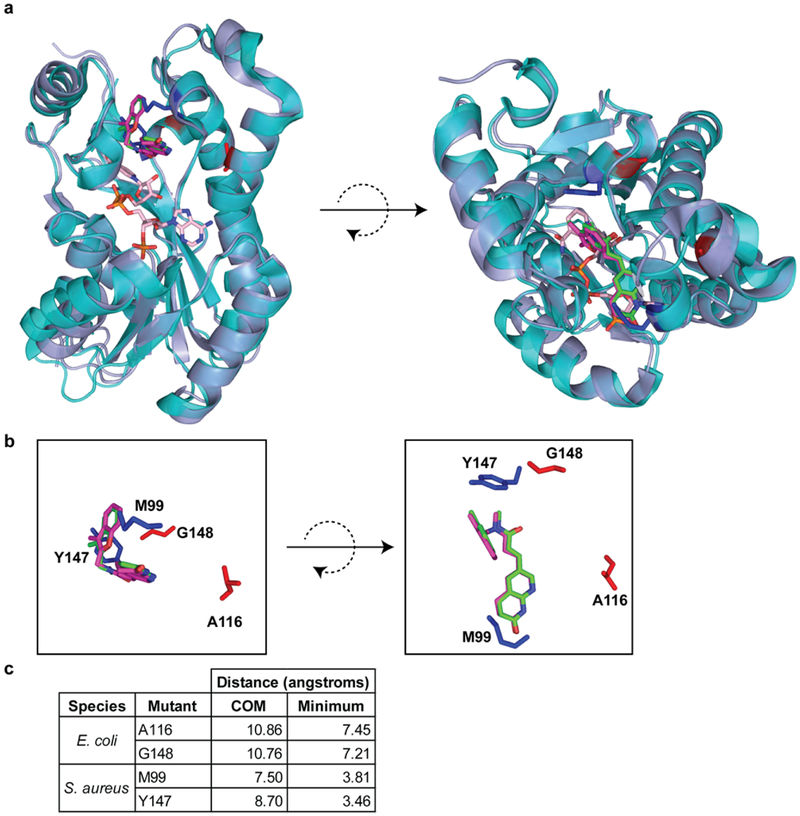
Extended Data Fig. 8. Location of mutated residues that confer resistance to Debio-1452 and Debio-1452-NH3.
a. Structural alignment of E. coli FabI (periwinkle, PDB 4JQC) and S. aureus FabI (turquoise, PDB 4FS3). Residues mutated in E. coli are highlighted in red. Residues mutated in S. aureus are highlighted in blue. Debio-1452 from E. coli structure is green and from S. aureus is magenta. NADPH from S. aureus structure is pink. b. Same poses as a but without cartoon representation of protein backbone and NADPH. c. Distances from mutation sites and Debio-1452, either between centers of mass (COM) or the minimum pairwise distance between atoms within mutated residues and Debio-1452.