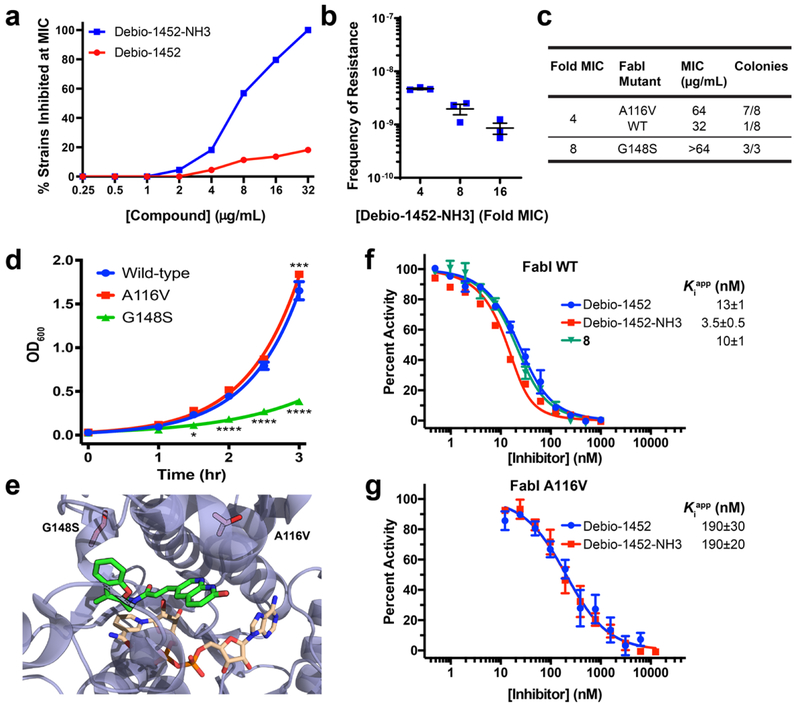
Fig. 2. Debio-1452-NH3 Mode of Action Studies.
a. Antimicrobial activity of Debio-1452 and Debio-1452-NH3 against a panel of Enterobacteriaceae clinical isolates. All experiments were performed in biological triplicate. b. Spontaneous frequency of E. coli MG1655 resistance to Debio-1452-NH3 as a function of concentration. Data shown represent the average of three independent experiments. Error bars represent standard error of the mean. c. Relative prevalence of FabI variants in E. coli resistant to Debio-1452-NH3. Resistant colonies of E. coli MG1655 were isolated following selection at 4- and 8-fold the MIC of Debio-1452-NH3. d. Fitness of E. coli isolates that harbor the FabI variant conferring resistance to Debio-1452-NH3 (either A116V or G148S) were evaluated relative to parental strain. Data points represent average of four independent experiments. Error bars represent standard error of the mean. Measurements were compared by repeated measure two-way ANOVA. Tukey’s multiple comparisons test was used to compare strains at each timepoint. Statistical significance from the wild-type strain (E. coli MG1655) is indicated with asterisks (* p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001). e. Location of E. coli FabI amino acid substitutions conferring resistance to Debio-1452-NH3 (red residues, A116V and G148S). PDB: 4JQC. f. In vitro inhibition of E. coli FabI and (g) the A116V FabI variant by Debio-1452 and derivatives. Initial reaction rates were used to calculate percent activity relative to inhibitor-free reaction. Dose-response curves were fit to Morrison’s quadratic to determine apparent Ki. Data is represented as average of three independent experiments. Error bars represent standard error of the mean.