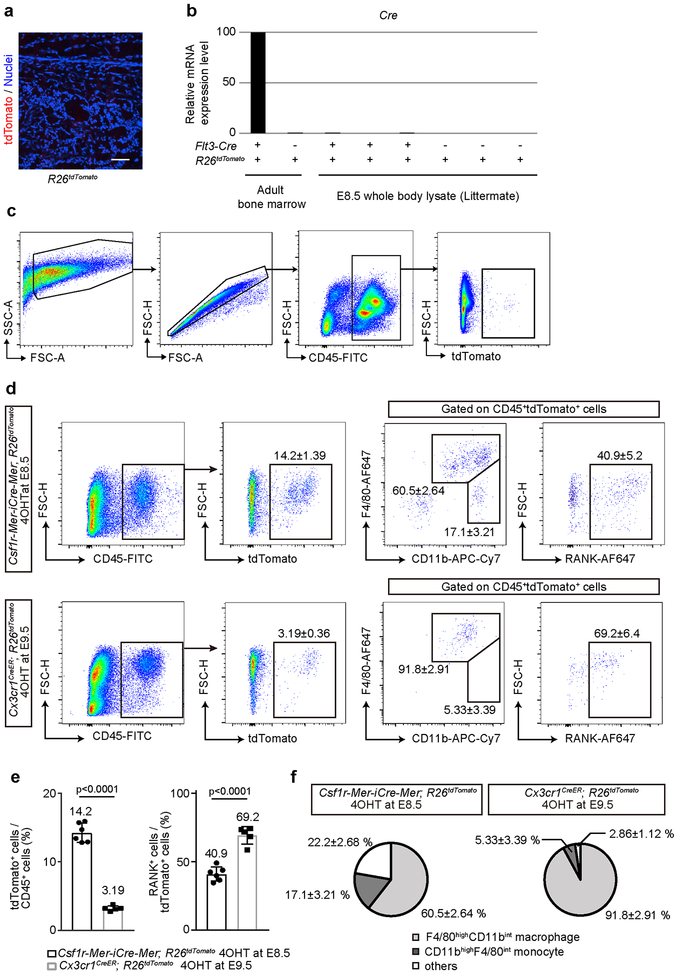
Extended Data Fig. 3.
Characterization of HSC- and EMP- derived cells. (a) Representative image of the femur of embryonic day (E) 17.5 R26tdTomato mice (Cre-negative littermate control of Flt3-Cre; R26tdTomato mice, n = 3 mice). Scale bar, 100 μm. (b) Relative mRNA expression levels of Cre were analyzed by quantitative PCR. RNA was isolated from adult bone marrow cells (BMCs) and E8.5 whole body. Flt3-Cre; R26tdTomato mice and their littermate Cre-negative control were used. (c) Gating strategy of CD45+tdTomato+ BMCs. (d) Flow cytometry analysis of tdTomato+ cells from whole-body cell suspension of E14.5 Csf1r-Mer-iCre-Mer; R26tdTomato mice labeled at E8.5 (n = 6 embryos) and Cx3cr1CreER; R26tdTomato mice labeled at E9.5 (n = 5 embryos). 4OHT, 4-hydroxytamoxifen. (e) Quantitative visualization of percentage of tdTomato+ and RANK+ cells from whole-body cell suspension of E14.5 Csf1r-Mer-iCre-Mer; R26tdTomato mice labeled at E8.5 (n = 6 embryos) and Cx3cr1CreER; R26tdTomato mice labeled at E9.5 (n = 5 embryos). Unpaired two-tailed t-test. Error bars denote means ± s.d. (f) Percentage of F4/80highCD11bint macrophage and CD11bhighF4/80int monocyte isolated from E14.5 whole body lysate of Csf1r-Mer-iCre-Mer; R26tdTomato mice labeled at E8.5 (n = 6 embryos) and Cx3cr1CreER; R26tdTomato mice labeled at E9.5 (n = 5 embryos). Statistics source data are provided in Source Data Extended Data Fig. 3.