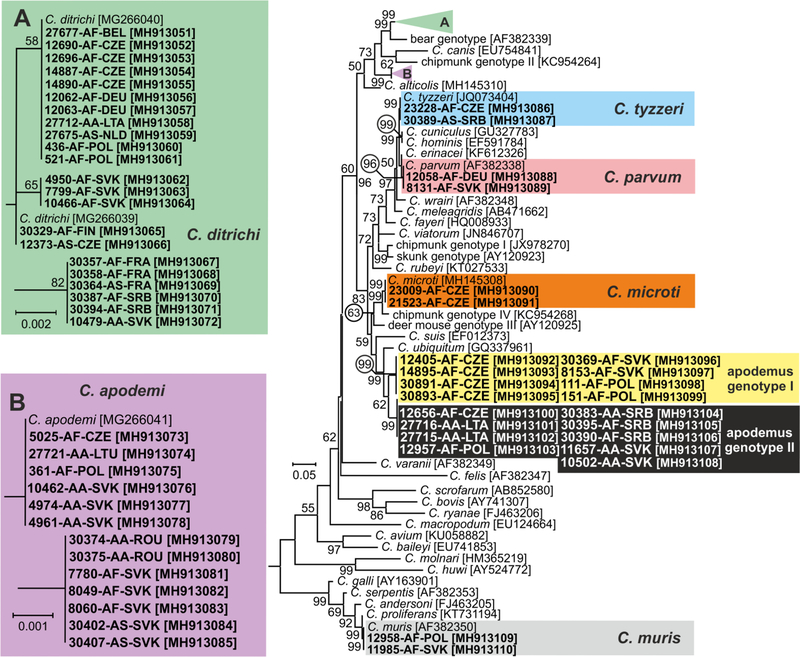
Fig. 2.
Maximum likelihood tree (−ln = 7878.45) based on partial sequences of gene coding actin of Cryptosporidium, including sequences obtained in this study (highlighted and bolded). The alignment contained 695 base positions in the final dataset. The General Time Reversible model was applied, using a discrete Gamma distribution and invariant sites. Numbers at the nodes represent the bootstrap values with more than 50% bootstrap support from 1000 pseudoreplicates. The branch length scale bar, indicating the number of substitutions per site, is given in the tree. Sequences from this study are identified by isolate number (e.g. 8131), host species (AA for Apodemus agrarius, AF for Apodemus flavicollis and AS for Apodemus sylvaticus) and region (BEL for Belgium, CZE for Czech Republic, FIN for Finland, FRA for France, DEU for Germany, LTA for Latvia, LTU for Lithuania, NLD for Nederland, POL for Poland, ROU for Romania, SRB for Serbia and SVK for Slovakia).