Fig 2. Dendrogram representing the genotype diversity and genetic relationships estimated by the cluster analysis of BOX-PCR fingerprints of diazotrophic/N-scavenging bacteria isolated from different sources and from soils under different management conditions.
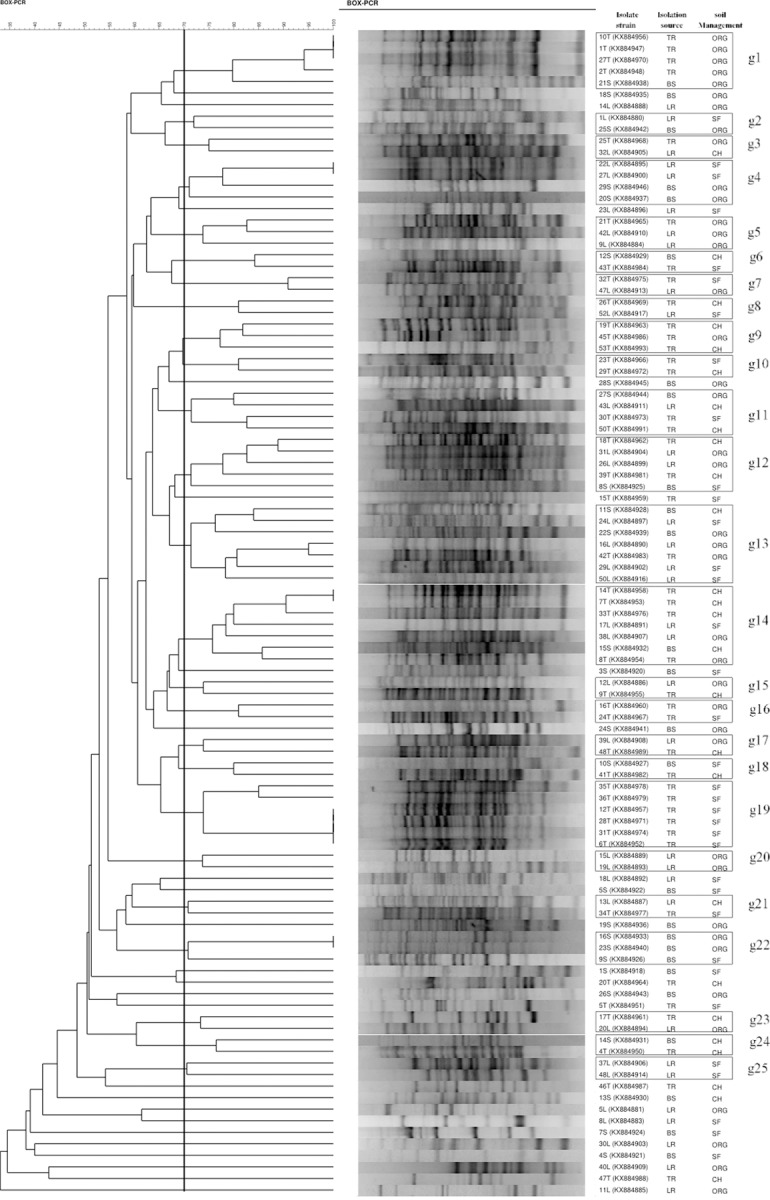
Dendogram was constructed using the Jaccard coefficient (2% tolerance in terms of band size) with the UPGMA algorithm. Isolation sources: BS (soil); LR (lulo unwashed roots), TR (tomato unwashed roots). Soil management conditions: CH (horticulture under conventional management); ORG (horticulture under organic management); SF (secondary forest with no agricultural use).
