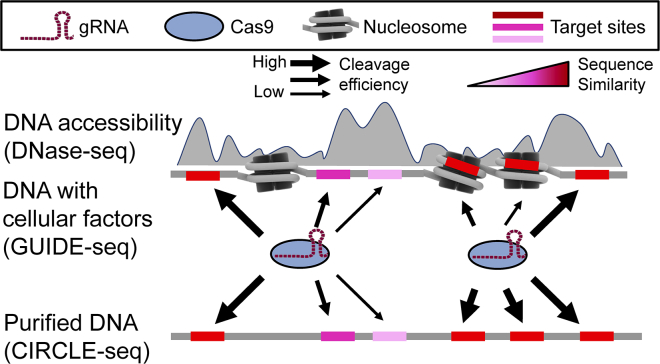
Figure 1.
The Schematic Diagram Demonstrates the Hypothesis that Presence of Cellular Factors in the GUIDE-Seq Experiment Reduces CRISPR-Induced Cleavage Efficiency
The CRISPR-induced cleavage efficiency could be negatively affected by the lack of DNA accessibility even with high sequence similarity between gRNA and target. The gray shade represents local DNA accessibility. The shade of potential target sites represents the sequence similarity of gRNA:target pairing. Note that the sequence similarity could be implied by any matrices that describe the sequence recognition pattern for the CRISPR-Cas9 system. The thickness of black arrow represents the level of CRISPR-induced cleavage efficiency. The gray shade behind the nucleosome represents the DNase I hypersensitivity detected in the DNase-seq assay system.