Table 1.
Examples of molecular structural motif identification of bile metabolites by COLMAR and pNMR MSMMDB queries
| Chemical shifts of input (1H, 13C) spin pairs | Motif | Hit that contains true motif returned by COLMAR MSMMDB (Hit rank, type of identified true motif, RMSD (ppm)) | Hit that contains true motif returned by pNMR MSMMDB (Hit rank, type of identified true motif, RMSD (ppm)) | True Metabolite |
|---|---|---|---|---|
| (4.120, 86.907) (4.342, 76.337) (4.213, 72.108) (3.802, 63.425) (3.918, 63.385) (5.902, 92.079) |
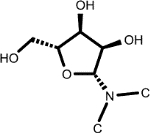 |
5-Methyluridine (1, 2nd shell, 0.16) | Beta-D-3-Ribofuranosyluric acid (2, 2nd shell, 2.08) | Uridine |
| (7.183, 133.486) (6.888, 118.557) |
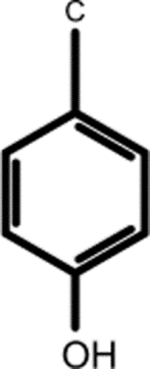 |
3-Chlorotyrosine (1, 2nd shell, 0.20) | N-(1-Deoxy-1-fructosyl)tyrosine (1, 2nd shell, 1.42) | L-tyrosine |
| (3.768, 74.784) (3.632, 65.208) (3.610, 73.570) |
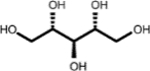 |
Adonitol (1, 2nd shell, 0.79) | Xylitol (1, 2nd shell, 1.80) | Xylitol |
| (1.788, 33.111) (2.996, 41.918) (4.135, 57.765) (1.710, 29.096) (1.398, 24.725) |
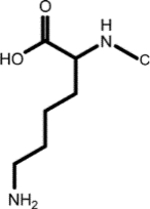 |
Biocytin (1, 1st shell, 1.29) | N6-L-homocysteinyl-N2-L-valyl-L-lysine (3, *2nd shell, 1.29) | Aspartyl-lysine |
| (3.966, 62.877) (3.234, 43.195) |
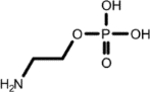 |
Ethanolamine (1, 1st shell, 1.63) | Phosphoethanolamine (1, 2nd shell, 1.44) | Phosphoethanolamine |
| (4.243, 68.598) (3.575, 63.141) (1.318, 22.145) |
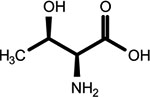 |
O-phosphothreonine (l, *2nd shell, 1.64) | Cyclic N(6)-threonylcarbamoyladenosine (1, 2nd shell, 1.83) | Threonine |
For these molecules, a partial 2nd shell MSM was identified, since in the true molecule a hydrogen terminates the 1st shell MSM.
