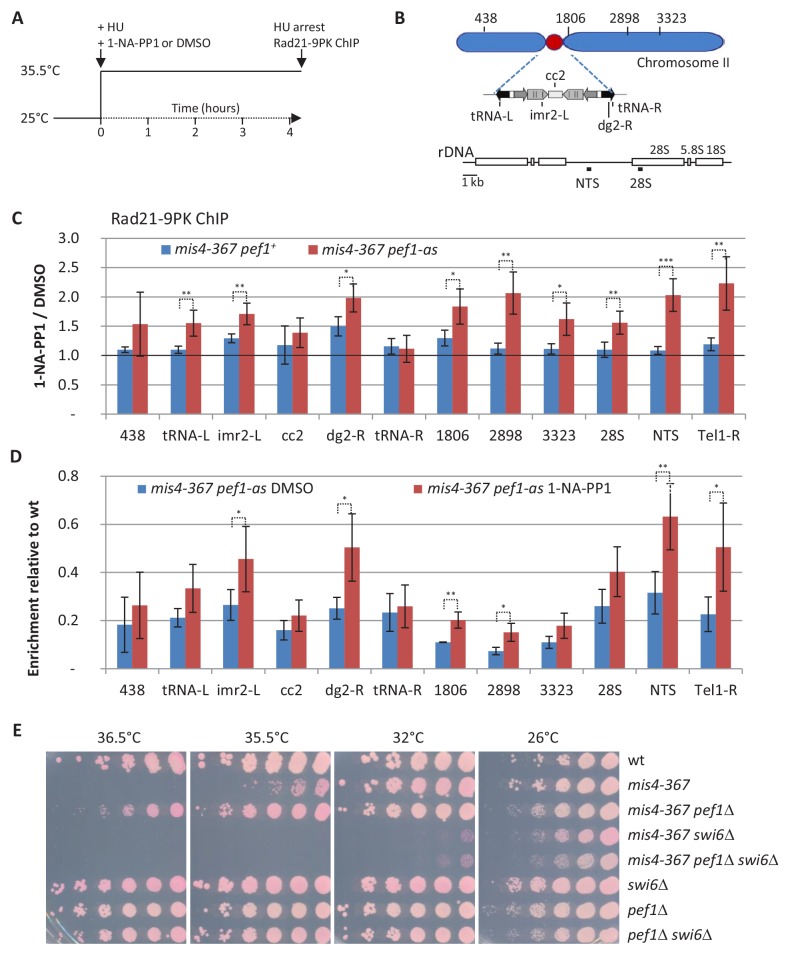
Figure 2. Inhibition of the CDK Pef1 in mis4-367 increased Rad21 binding to S phase chromosomes.
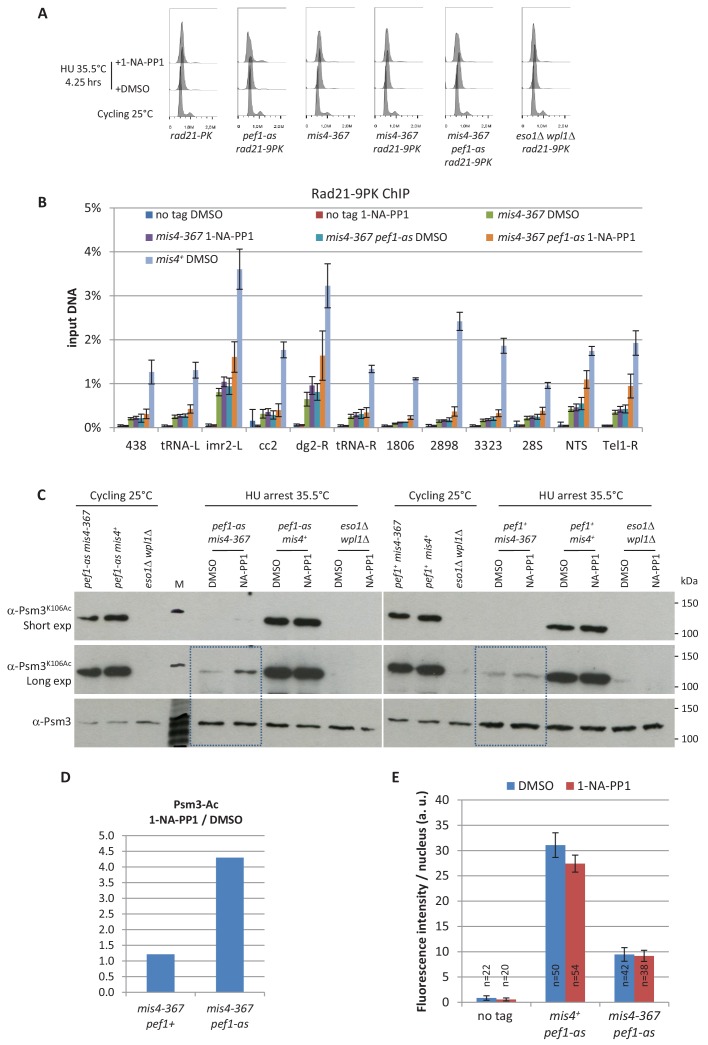
(A) Scheme of the experiment. Hydroxyurea (HU) was added to 12 mM at the time of the temperature shift along with 1-NA-PP1 or solvent alone (DMSO). Cells were collected after 4.25 hr. The S phase arrest was confirmed by DNA content analysis (Figure 2—figure supplement 1). (B) Schematics showing the loci analyzed by ChIP-qPCR. (C) The effect of 1-NA-PP1 treatment on Pef1-as is shown by the ratio 1-NA-PP1/DMSO (red) for each site analyzed. The ratios in a pef1+ background (blue) estimate the off target effects of the inhibitor. Ratios were calculated from the ChIP data shown in Figure 2—figure supplement 1. Bars indicate mean ± SD from four ratios. ***p≤0.001, **p≤0.01, *p≤0.05, by two-tailed, unpaired t-test with 95% confidence interval (Figure 2—source data 1). (D) Rad21 binding relative to wild-type. The ratios highlight the recovery of Rad21 binding upon inhibition of the CDK relative to wild-type levels. Ratios were calculated from the data shown in Figure 2—figure supplement 1. Bars indicate mean ± SD from four ratios. **p≤0.01, *p≤0.05, by two-tailed, unpaired t-test with 95% confidence interval (Figure 2—source data 1). (E) Cell growth assay showing that pef1Δ does not suppress mis4-367 thermosensitive phenotype in the absence of the swi6 gene.