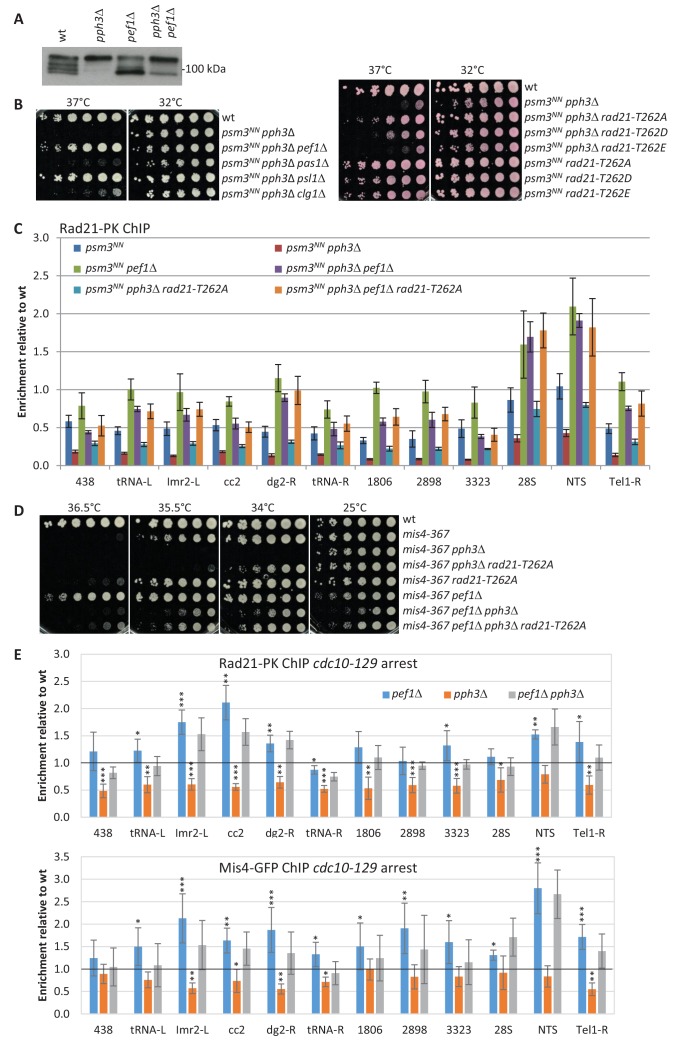
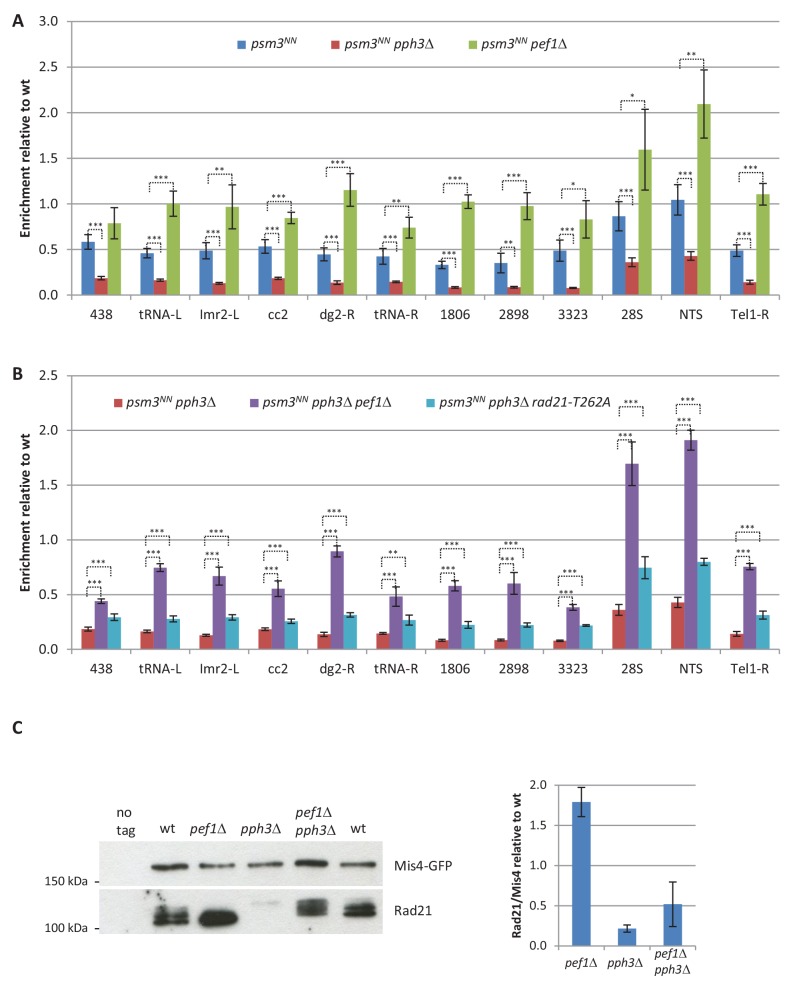
Figure 7. Pef1 and PP4 oppose each other for regulating Rad21 binding to chromosomes.
(A) Western blot analysis of total protein extracts from cycling cells probed with anti-Rad21 antibodies. (B) Cell growth assays showing that the ts growth defect of psm3NN pph3Δ is efficiently rescued by pef1 and psl1 deletion mutants (left) and rad21-T262A (right). (C) Rad21-ChIP after one complete cell cycle at 36°C. Data are expressed relative to wild-type. Bars indicate mean ± SD from four ratios. t-tests are shown in Figure 7—figure supplement 1 and Figure 7—source data 1. (D) Cell growth assays showing that pef1Δ, rad21-T262A and pph3Δ display opposite genetic interactions with mis4-367. (E) Rad21 and Mis4 ChIP relative to wild-type in cdc10-129-arrested cells. Bars indicate mean ± SD from four ratios (Figure 4—source data 1).