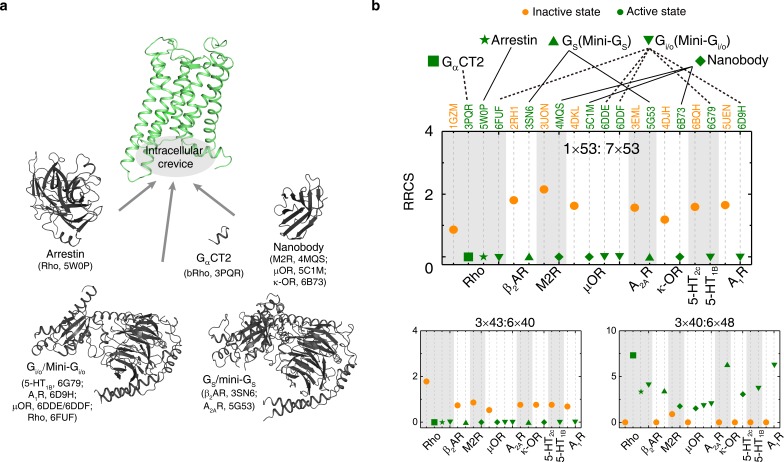
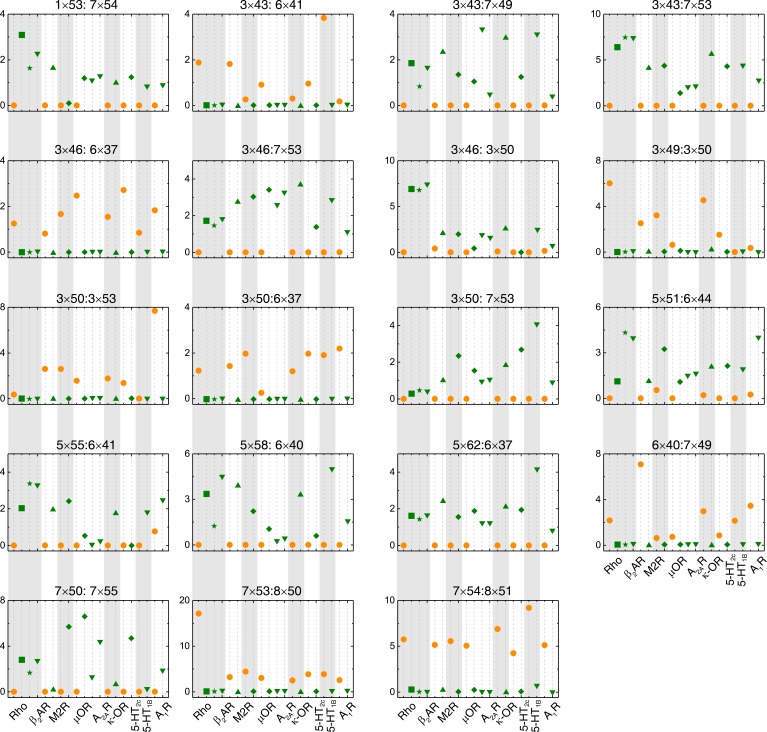
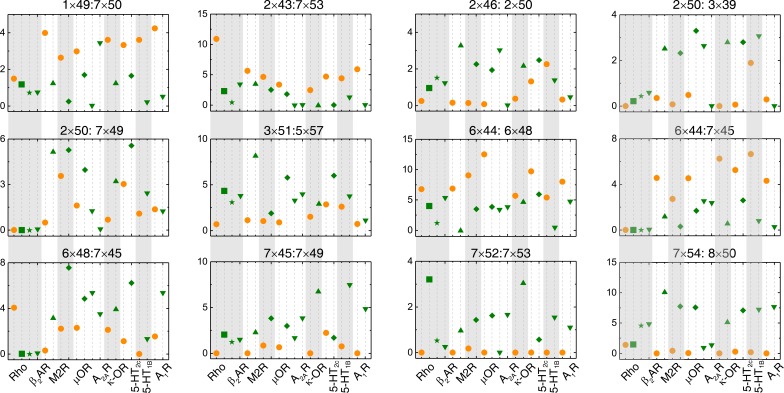
Figure 4. The common activation mechanism is the shared portion of various downstream pathways of different class A GPCRs.
(a) Intracellular binding partners used in the active state structures. (b) Comparison of RRCS for active (green) and inactive (orange) states of eight receptors with different intracellular binding partners, including four recently solved cryo-EM structures of Gi/o-bound receptors (5-HT1B receptor, rhodopsin, A1R and µOR) (Tsai et al., 2018; García-Nafría et al., 2018; Kang et al., 2018; Koehl et al., 2018; Draper-Joyce et al., 2018) whose resolutions were low (usually ≥3.8 Å for the GPCR part). Nevertheless, almost all conserved residue rearrangements in the pathway can be observed from them. Three of 34 residues pairs were shown here, see Figure 4—figure supplements 1 and 2 for the remaining 31 residue pairs.