Fig. 2. Long-read transcriptomics.
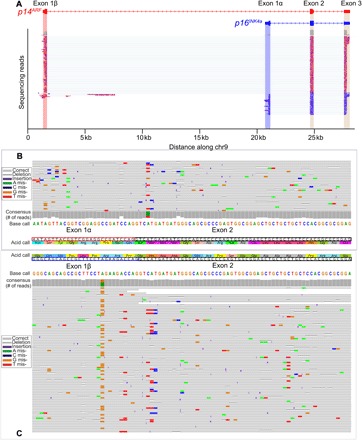
(A) Direct RNA sequencing reads from GM12878 are shown mapped to the CDKN2B genomic locus. Plotted by Integrated Genomics Viewer, individual RNA reads aligned against the reference human genome (hg38) in this region are shown with the reads matching to the p14ARF isoform colored red and the p16INK4a isoform colored blue. Note that exons 2 and 3 are in common, while exon 1α is specific to p16 and exon 1β is specific to p14. (B and C) Zoomed-in regions are shown with predicted translations at the 5′ exon 2 boundary. Reads are aligned against the transcriptome (Gencode v27). Integrative Genomics Viewer reads aligned against the transcriptome (Gencode v27) are shown with p16 on the top and p14 on the bottom. Although insertions/deletions (indels) and mismatches with the reference are observed, the consensus agrees 99% of the time with the reference. The translated codons are shown between the aligned reads; note that although exon 2 is the same RNA sequence in both, the resulting protein is completely different because of the shifted reading frame from the splice variation.
