Fig. 1. The paradox of chromatin folding and the basic ideas of SRRW.
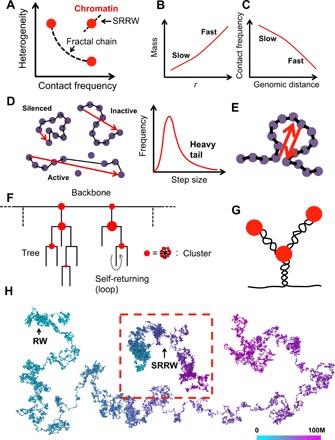
(A) Chromatin structure cannot be explained by a fractal chain that has anticorrelated contact frequency and density heterogeneity. SRRW is developed to unify these two properties. (B) Schematic representation of the mass scaling behavior of chromatin. (C) Schematic representation of the contact scaling behavior of chromatin (<10 Mb), which is counterintuitive given the mass scaling behavior. (D) Coarse-graining diverse epigenetic states at the nanoscale into a wide distribution of step sizes. One step approximately maps to 10 nucleosomes or 2-kb DNA. The balls represent histones, and the lines represent DNA. The arrows represent the coarse-grained steps in SRRW. (E) Self-returning as a coarse-grained representation of looping (in a broad sense including supercoiling and clustering). (F) SRRW’s topological architecture featuring random trees connected by a backbone. Tree nodes are formed by frequent self-returning of short steps. (G) One possible realization of tree structures is by combining nanoclusters with nested supercoils or loops. (H) Comparison between a free SRRW (highlighted in the red box) and a free RW, both of 50,000 steps (100 Mb)
