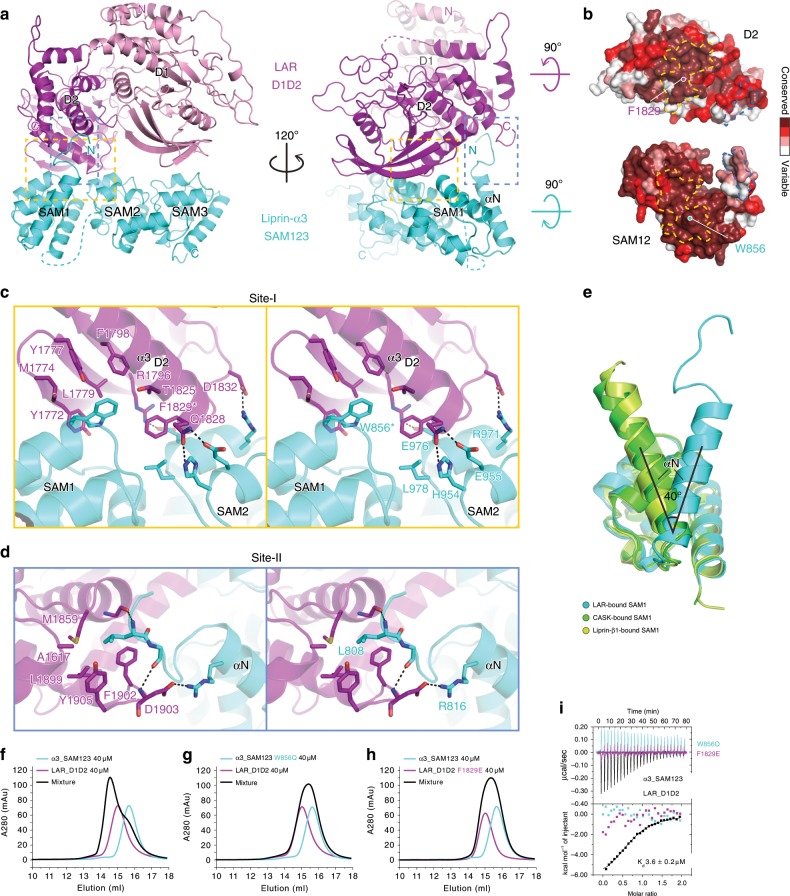
Fig. 2. Structural characterization of the liprin-α3/LAR interaction.
a Ribbon representations of the α3_SAM123/LAR-D1D2 complex structure. The two binding sites were highlighted by dashed boxes. b Surface representations showing the high conservation of the two binding sites. c, d Stereoview of the atomic details of the two binding sites between α3_SAM123 and LAR-D1D2, corresponding to the boxed regions shown in (a) with the same color. W856 in liprin-α3 and F1829 in LAR, indicated by asterisks, play the key role in the liprin-α3/LAR interaction. Hydrogen bonds and salt bridges are indicated by dashed lines. e Structural alignment of the SAM1 domains from the complex structures of α3_SAM123/LAR-D1D2, α2_SAM123/CASK_CaMK (PDB ID: 3TAC), and α2_SAM123/ β1_SAM123 (PDB ID: 3TAD) showing the orientation change of the αN-helix. f–h Analytical gel filtration analysis showing that either the W856Q mutation in liprin-α3 or the F1829 mutation in LAR disrupts the liprin-α3/LAR interaction. i ITC-based measurement of the binding of α3_SAM123 or its W856Q mutant to LAR_D1D2 and the binding of α3_SAM123 to the F1829E mutant of LAR_D1D2.