Abstract
Arabidopsis SIZ1 encodes a SUMO E3 ligase to regulate abiotic and biotic stress responses. Among SIZ1 or mammalian PIAS orthologs, plant SIZ1 proteins contain the plant homeodomain (PHD) finger, a C4HC3 zinc finger. Here, we investigated the importance of PHD of Arabidopsis SIZ1. The ProSIZ1::SIZ1(ΔPHD):GFP was unable to complement growth retardation, ABA hypersensitivity, and the cold-sensitive phenotype of the siz1 mutant, but ProSIZ1::SIZ1:GFP could. Substitution of C162S in the PHD finger was unable to complement the siz1 mutation. Tri-methylated histone H3K4 (H3K4me3) was recognized by PHD, not by PHD(C162S). WRKY70 was up-regulated in the siz1-2 mutant and H3K4me3 accumulated at high levels in the WRKY70 promoter. PHD interacts with ATX, which mediates methylation of histone, probably leading to suppression of ATX’s function. These results suggest that the PHD finger of SIZ1 is important for recognition of the histone code and is required for SIZ1 function and transcriptional suppression.
Subject terms: Plant sciences, Plant stress responses, Abiotic
Kenji Miura et al. investigate the role of the plant homeodomain (PHD) finger of the Arabidopsis SIZ1 protein. They show that the PHD finger is involved in hormone response and temperature sensitivity, and plays an important role in H3K4 methylation, thereby affecting recognition of histone code and transcriptional suppression.
Introduction
Sumoylation, the covalent attachment of small ubiquitin-like modifier (SUMO) to other proteins, is a post-translational modification that controls protein function, activity, localization, and turnover in eukaryotes1,2. In plants, sumoylation is involved in the response to abiotic and biotic stresses, such as cold, salt, and drought stresses, and innate immunity3–8. Furthermore, sumoylation regulates signaling pathways for plant hormones, including abscisic acid (ABA) and salicylic acid7,9–12. Similar to ubiquitin, three enzymes, E1, E2, and E3, are required for the attachment of SUMO to other proteins2. E1, the heterodimeric SUMO-activating enzyme, which is composed of the SAE1 and SAE2 subunits, binds to SUMO via a high-energy thioester linkage13,14. Activated SUMO is transferred to E2, SUMO-conjugating enzyme 1 (SCE1) via transesterification, and then conjugated to substrate proteins, assisted by E3, SUMO ligase15,16, via an isopeptide bond between the C-terminal glycine of SUMO and specific lysine(s) within the target. SUMO is often conjugated to lysine in the conserved ΨKXE/D (Ψ, a large hydrophobic amino acid; K, lysine; X, any amino acid; E, glutamate; and D, aspartate) motifs. At the molecular level, sumoylation alters the function of the targets, including changes in their intracellular localization, activity, and interaction with other proteins17. Previous proteomic studies have identified over 1000 SUMO1/2 targets in Arabidopsis18,19. Most of these targets are nuclear-localized proteins and these proteins have functions related to DNA modification, chromatin assembly, transcription factors, coactivators/repressors, and abiotic and biotic stress responses19.
In Arabidopsis, four SUMO E3 ligases have been identified; SAP and MIZ1 domain-containing ligase1 (SIZ1)15, methyl methansulfonate-sensitive21 (MMS21)/high ploidy2 (HPY2)16,20, and protein inhibitors of activated STATs-like1 (PIAL1) and PIAL221. All ligases contains the SP-RING (Siz/PIAS-RING) domain, which functions for SUMO E3 ligase activity. The siz1-2 hpy2-1 double mutation causes embryonic lethality22, indicating that these SUMO E3 ligases play important roles in sumoylation in Arabidopsis but they have distinct role in development22. Among the > 1000 SUMO targets in Arabidopsis, few could be assigned to MMS21, whereas numerous targets could be assigned to SIZ119, suggesting that MMS21 modifies a small number of proteins and that both SUMO and SIZ1 are crucial regulators of chromatin function and transcription. PIAL1 and PIAL2 function as E4-type SUMO ligases to promote SUMO chain formation and are involved in salt and osmotic stress responses23. Among these four SUMO E3 ligases, SIZ1 has high similarity to yeast Siz and animal PIAS orthologs24, which contain SAP and SP-RING domains and PINIT (for Pro-Ile-Asn-Ile-Thr) and SXS (Ser-any amino acid-Ser) motifs15,25. However, only plant SIZ proteins contain a PHD (plant homeodomain) finger, which is a C4HC3 (four cysteines, one histidine, and three cysteines) zinc-finger-like motif. The PHD finger proteins of ING2 and bromodomain and PHD finger transcription factor (BPTF) recognize trimethylated Lys4 of histone H3 (H3K4me3)26,27. Thus, it is thought that PHDs read the epigenetic code28. Investigation of the PHD finger of rice SIZ1 bound to methylated histone H3 by NMR revealed that OsSIZ1-PHD recognized both dimethylated Arg2 and trimethylated Lys4 of histone H329. The PHD finger of Arabidopsis SIZ1 binds to AtSCE1 and is required for sumoylation of GTE3, a bromodomain protein, together with SP-RING domain, suggesting that PHD and SP-RING contribute to SUMO E3 ligase function30. Although the PHD finger seems to be important for biochemical function, a point mutation in the PHD of Arabidopsis SIZ1 complemented several siz1-2 phenotypes, such as plant growth retardation, thermosensitive seed germination, and hypersensitivity to ABA-induced inhibition of cotyledon expansion25. Conversely, point-mutated SP-RING was unable to complement the siz1-2 phenotype25.
To confirm biological importance of the PHD finger in SIZ1, we transformed ProSIZ1::SIZ1(ΔPHD):GFP into the siz1-2 mutant. Although ProSIZ1::SIZ1:GFP was able to complement the siz1 mutation31, ProSIZ1::SIZ1(ΔPHD):GFP was not, suggesting the biological importance of the PHD finger of SIZ1. In addition, ProSIZ1::SIZ1(C162S):GFP was not able to complement it. The biochemical function of PHD is the preferential recognition of histone H3K4me3. Substitution of C162S in the PHD finger prevented recognition of histone H3K4me3, probably preventing complementation of the siz1-2 mutation. Histone H3K4me3 is enriched with transcriptionally active promoters32. In human cells, recognition of H3K4me3 by ING2-PHD stabilizes the mSin3a–HDAC1 complex to repress active genes in response to DNA damage33. Because H3K4me3 was highly accumulated in the promoter of WRKY70 in the siz1-2 mutant, SIZ1 was suggested to repress active WYKY70 gene expression via recognition of H3K4me3 by the PHD finger. ATX proteins methylate histone H3K434,35. The PHD finger also interacts with ATX proteins. It is likely that PHD suppresses the methylation function of ATX proteins. In this article, we demonstrate importance of the PHD finger of SIZ1 on recognition of histone code and transcriptional suppression.
Results
PHD finger is important for SIZ1 function
Plant SUMO E3 ligases, SIZs, contain several motifs and domains, such as SAP (Scaffold attachment factor A/B/acinus/PIAS), PHD, PINIT, SP-RING, SXS, and NLS15,25. Among them, the PHD finger is a unique domain of plant SIZ proteins, whereas SIZ/PIAS proteins in yeast and animals contain no PHD finger. In vitro analysis revealed that the PHD and SP-RING domains of Arabidopsis AtSIZ1 are required for binding to the AtSCE1, the SUMO E2-conjugating enzyme, and for sumoylation30. Thus, the PHD finger is assumed to be important for function of AtSIZ1. However, substitution of C134 to tyrosine in the PHD of AtSIZ1 was able to complement phenotypes of the siz1-2 mutant, such as dwarf-like, thermosensitivity of seed germination, and ABA hypersensitivity25. Expression of AtSIZ1(C134Y) in siz1-2 resulted in abnormal hypocotyl elongation in response to sugar and light, whereas the siz1-2 mutant did not exhibit such a phenotype25.
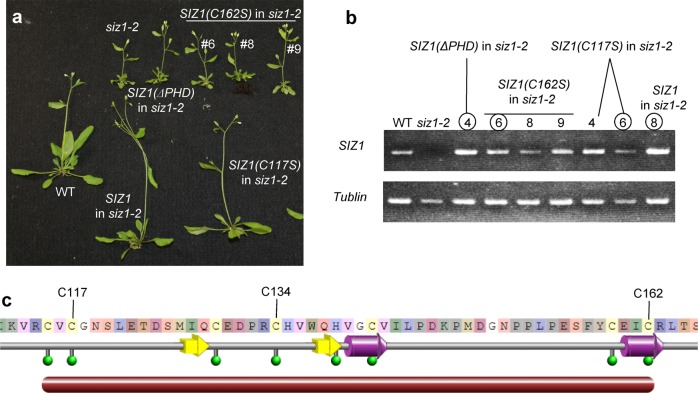
To confirm whether the PHD finger is important for SIZ1 function, ProSIZ1::SIZ1(ΔPHD):GFP was expressed in the siz1-2 mutant to complement the dwarf-like phenotype. Expression of ProSIZ1::SIZ1(ΔPHD):GFP was unable to complement the dwarf-like phenotype of the siz1-2 mutant, whereas ProSIZ1::SIZ1:GFP was (Fig. 1a). The expression of SIZ1(ΔPHD):GFP was confirmed by RT-PCR (Fig. 1b). The results suggest that PHD is important for complementing the dwarf-like phenotype of the siz1-2 mutant. Then, a substitution was introduced in ProSIZ1::SIZ1:GFP and transformed into the siz1-2 mutant. Because the PHD finger is a C4HC3 zinc-finger domain and cysteines and histidine are required for binding to zinc36, ProSIZ1::SIZ1(C117S):GFP or ProSIZ1::SIZ1(C162S):GFP was expressed in the siz1-2 mutant. Expression of SIZ1(C117S), as well as SIZ1(C134Y), was able to complement the dwarf-like phenotype25, but SIZ1(C162S) was not (Fig. 1a). Expression of SIZ1 variants was confirmed by RT-PCR (Fig. 1b). These results indicate that C162 in PHD is important for complementation of the siz1-2 mutation. The amino-acid sequence of the PHD finger in SIZ1 demonstrated that C162 is in an α-helix, whereas C117 and C134 are not (Fig. 1c). It is probable that substitution of C162 to serine severely affects SIZ1 function.
Fig. 1. Mutation in the plant homeodomain (PHD) finger of SIZ1 was unable to complement the dwarf-like phenotype of the siz1-2 mutant.
a ProSIZ1::SIZ1:GFP or its variants were transformed into the siz1-2 mutant. Six-week-old siz1-2 mutant exhibited a dwarf-like phenotype, as previously described39. Introduction of ProSIZ1::SIZ1:GFP or ProSIZ1::SIZ1(C117S):GFP complemented the dwarf-like phenotype of siz1-2; however, introduction of ProSIZ1::SIZ1(C162S):GFP or ProSIZ1::SIZ1(ΔPHD):GFP was unable to complement the dwarf-like phenotype of siz1-2. b Expression of SIZ1 in each plant. SIZ1 was not detected in the siz1-2 mutant. Conversely, SIZ1 transcripts were detected in other plants. c Amino-acid sequence and structural information of the PHD finger in SIZ1. Green circles indicate zinc-binding residues. Purple and yellow arrows indicate α-helices and β-sheets, respectively. The illustration was downloaded from Protein Data Bank Japan (PDBj, https://pdbj.org/mine/structural_details/1wew).
Next, we examined whether other phenotypes of the siz1-2 mutant were complemented by expression of SIZ1 variants. The siz1-2 mutant exhibited ABA hypersensitivity for primary root growth, compared with wild-type seedlings (Fig. 2a and b)37. Expression of SIZ1:GFP or SIZ1(C117S):GFP suppressed the ABA hypersensitivity of siz1-2 seedlings, whereas expression of SIZ1(ΔPHD):GFP or SIZ1(C162S):GFP did not (Fig. 2 and Supplementary Fig. 1). Furthermore, expression of SIZ1:GFP or SIZ1(C117S):GFP complemented the cold sensitivity and drought tolerance of siz1-2 plants, whereas expression of SIZ1(ΔPHD):GFP or SIZ1(C162S):GFP in the siz1-2 mutant still exhibited cold sensitivity (Fig. 3 and Supplementary Fig. 2) and drought tolerance (Fig. 4 and Supplementary Fig. 3), indicating that the PHD finger is required for SIZ1 function in response to ABA, cold stress, and drought stress.
Fig. 2. Mutation in the PHD finger of SIZ1 was unable to complement ABA inhibition of primary root growth in seedlings of the siz1 mutant.
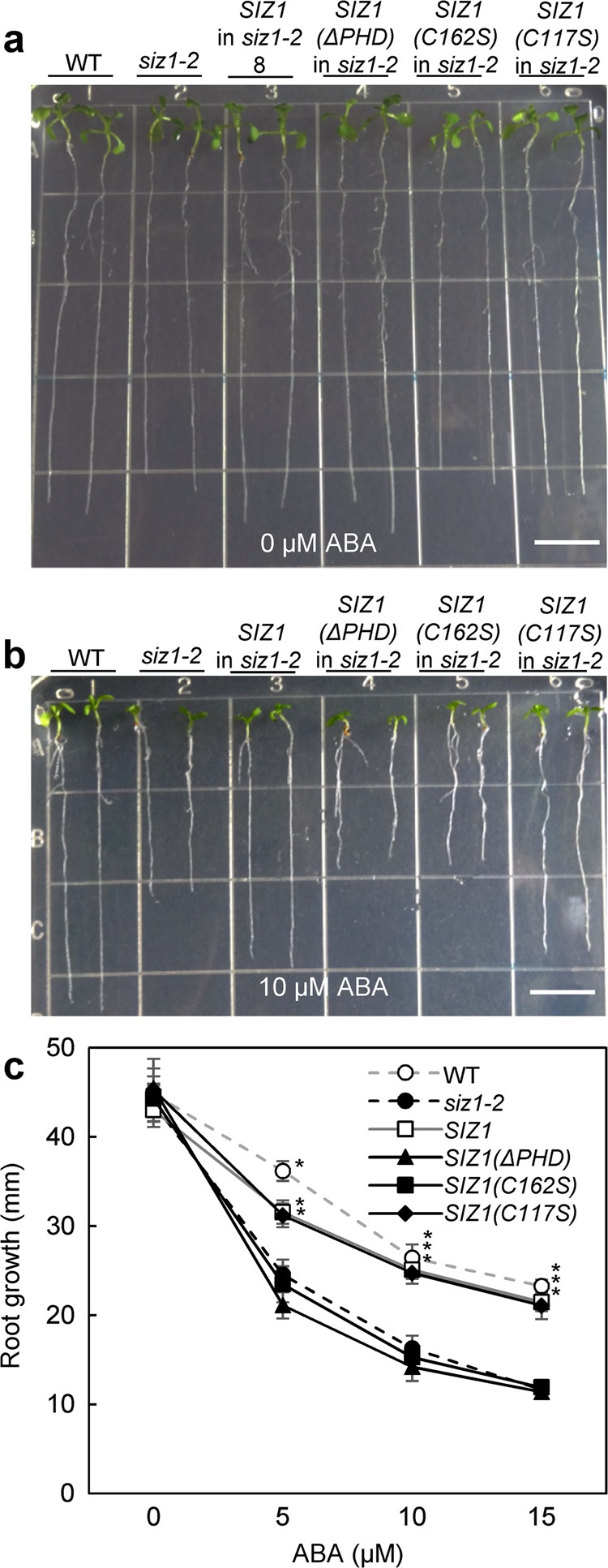
Three-day-old seedlings were transferred onto basal media containing 0 (a) or 10 µm ABA (b). The bar indicates 1 cm length. c Root growth values expressed as mean ± standard deviation (SD; n ≥ 15 biologically independent seedlings). Asterisk indicates a significant difference from the siz1 plants (p < 0.05) as determined by one-way ANOVA followed by the Tukey–Kramer test.
Fig. 3. Cold sensitivity caused by the siz1-2 mutation was not recovered by SIZ1 with mutation in the PHD finger.
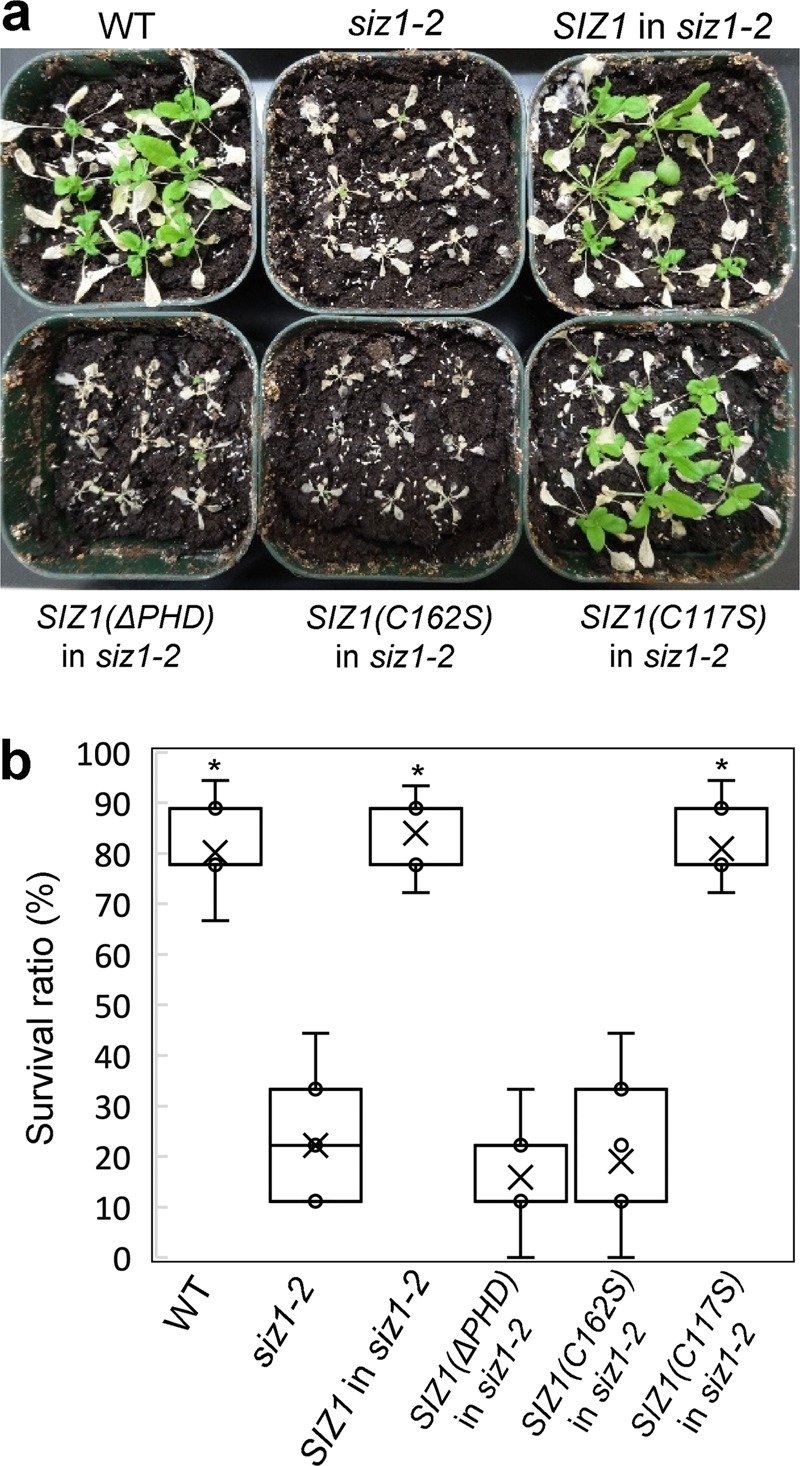
a Three-week-old plants were incubated at 4 °C for 1 week for cold acclimation. Cold-acclimated plants were exposed for 4 h at − 6 °C. Following freezing treatment, plants were incubated at 24 °C for 1 week. Photographs are representative of WT, siz1-2, and siz1-2 transformed with SIZ1 variants. b Survival was determined in 27 plants after freezing treatment at − 6 °C. Data are the mean ± SD calculated from four or more independent experiments. Asterisks indicate a statistical difference from the siz1-2 plants (p < 0.05) as determined by Student’s t test.
Fig. 4. Drought-tolerant phenotype of the siz1-2 mutant is not recovered by SIZ1 with mutation in the PHD finger.
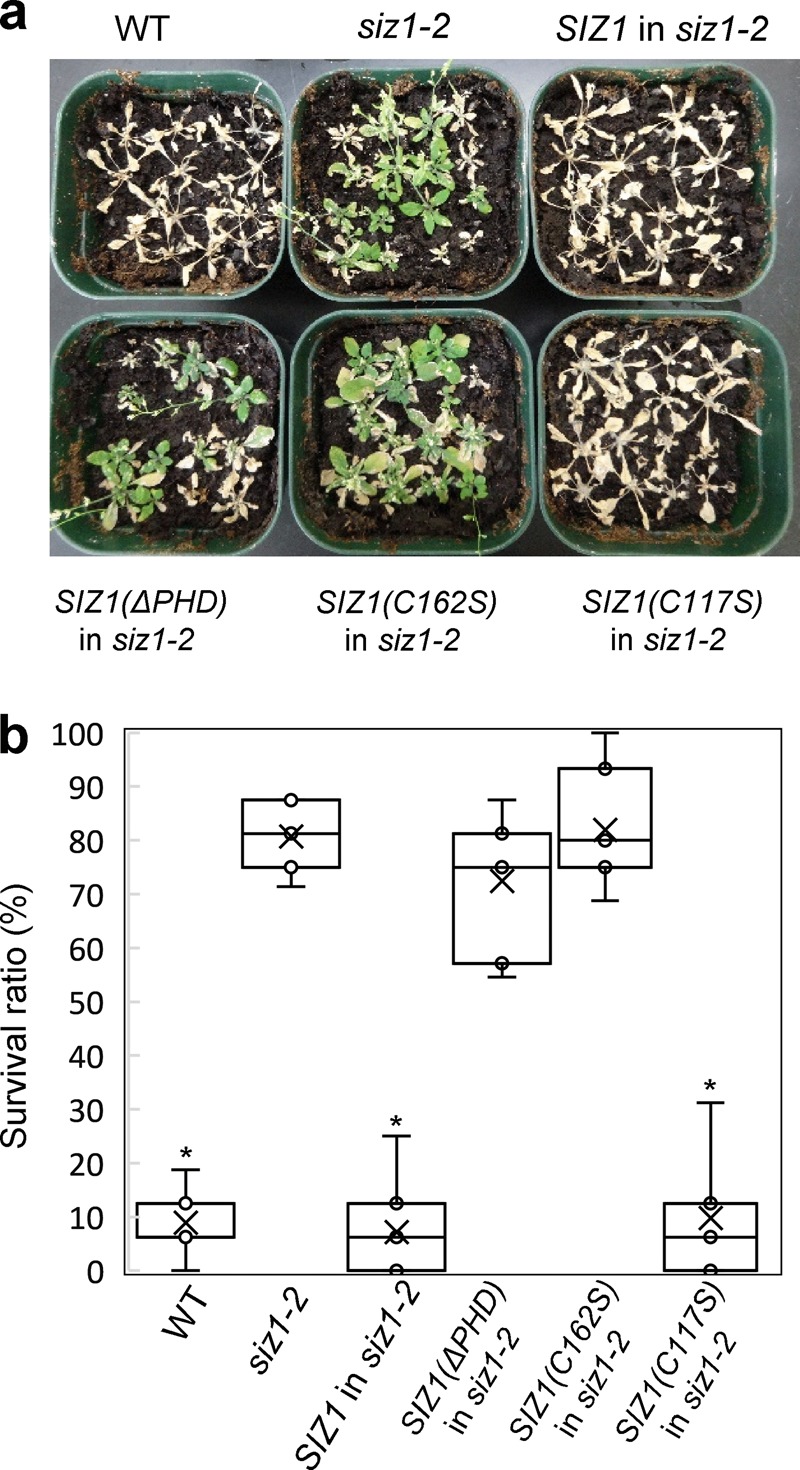
a Watering was resumed, and plants were incubated for 1 week after the 2-week drought treatment. b The survival ratio was determined for 16 plants after drought treatment. Data are mean ± SD from three independent experiments. Asterisks indicate a statistical difference from the siz1-2 plants (p < 0.05) as determined by Student’s t test. The data are a representative experiment from three independent experiments.
PHD recognizes trimethylated histone H3K4
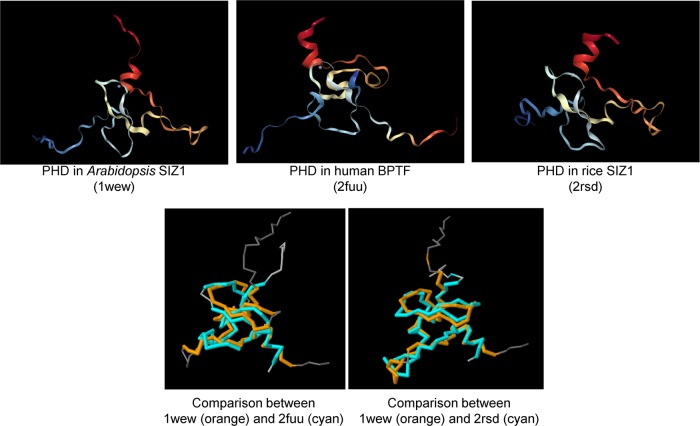
To elucidate the function of the SIZ1 PHD finger, its structure was compared with that in human and rice (Fig. 5). The structures of SIZ1 PHD (1wew), human BPTF PHD (2fuu), and rice SIZ1 PHD (2rsd) have been registered in the PDB database. The PHD finger of human BPTF recognizes histone H3K4me327. Furthermore, the PHD finger of rice SIZ1 interacts with histone H3K4me329. Because the structure of the PHD finger of SIZ1 is similar to that of BPTF and that of rice SIZ1 (Fig. 5), it is assumed that the PHD finger of SIZ1 recognizes histone H3K4me3.
Fig. 5. Comparison of the 3-D structure of the PHD finger in Arabidopsis SIZ1 (PDB ID: 1wew) and human BPTF (2fuu) or the PHD finger in rice SIZ1 (PDB ID: 2rsd).
The comparison was performed in PDB (http://www.rcsb.org/pdb/workbench/workbench.do).
The GST-PHD protein was incubated with several types of biotinylated histone H3. Histone H3 was pulled down with streptavidin beads and GST-PHD was detected with an anti-GST antibody. Based on the results of the pull-down assay, PHD interacted weakly with histone H3K4me2 and strongly with H3K4me3 (Fig. 6a). No interaction was detected when histone H3K9me1, H3K4me2, H3K9me3, H3K27me1, H3K427me3, or H3K36me1 was used (Fig. 6a, b). Next, GST-PHD(C162S) or GST-PHD(C117S) was prepared to confirm whether base substitution influenced the binding activity of PHD. GST-PHD(C162S) was not found to interact with histone H3K4me3 (Fig. 6c), suggesting that disrupting binding with histone H3K4me3 by substitution of cysteine to serine affects the function of SIZ1, as SIZ1(C162S):GFP was unable to complement the siz1-2 mutation. Conversely, GST-PHD(C117S) was able to bind to histone H3K4me3 (Fig. 6c); thus, the siz1-2 mutation was complemented by SIZ1(C117S):GFP. Because siz1-2 plants harboring SIZ1(C117S):GFP looked similar to wild-type plants, the binding activity of PHD(C117S) to histone H3 (Fig. 6c) may not affect the phenotype of the siz1-2 mutant harboring SIZ1(C117S):GFP. And difference of binding activity of PHD or PHD(C117S) to histone H3K4me3 may not be effective for complementation. For negative control, interaction between GST and histone H3 or H3K4me3 was examined (Fig. 6d).
Fig. 6. The PHD finger of SIZ1 recognizes trimethylated histone H3K4me3.
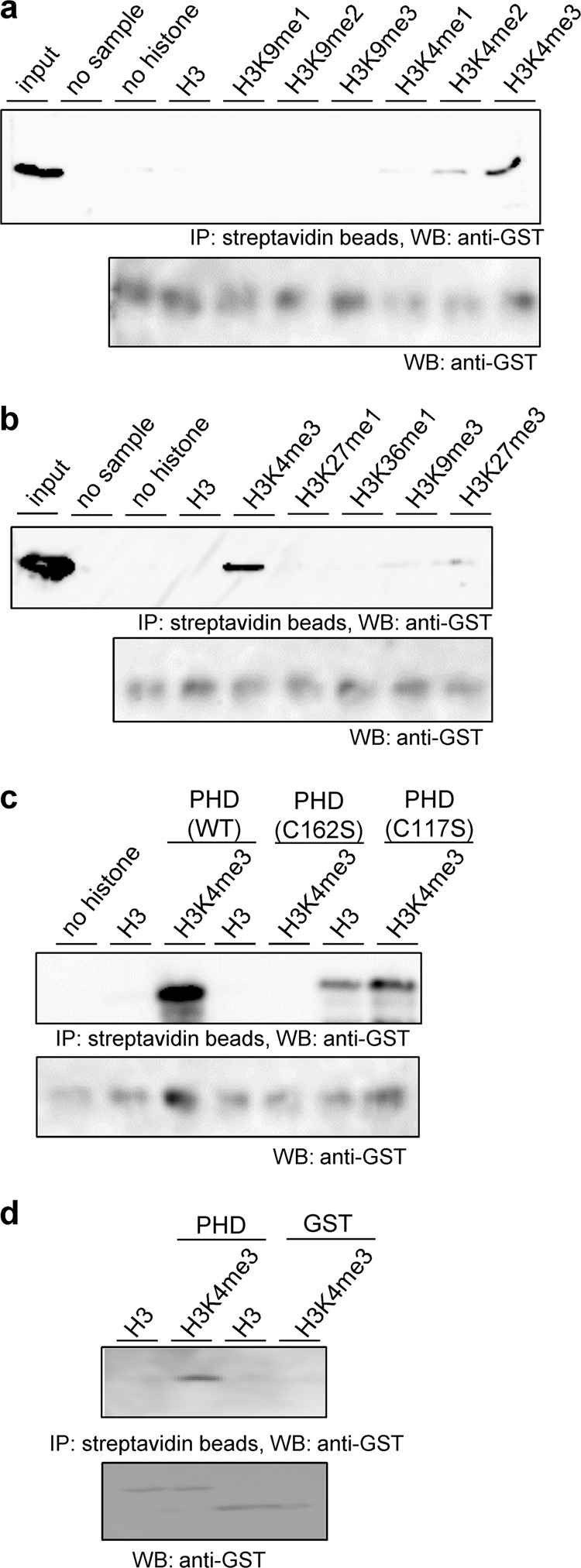
The PHD finger was inserted into pGEX5X-1 to produce a GST-PHD fusion protein. GST-PHD fusion proteins were incubated with histone H3 or methylated histone H3 fused with biotin. Pull-down of biotin-histone H3 was performed with streptavidin beads. Then, the proteins were detected by immunoblot analysis with anti-GST. a Histone H3 or methylated histone H3K4 or H3K9 were used for pull-down experiments. b Pull-down analysis was performed with several types of methylated histone H3. c The interaction between GST-PHD variants and histone H3K4me3 was investigated. GST-PHD, GST-PHD(C162S), and GST-PHD(C117S) were prepared and pull-down experiments were performed. d For negative control, pull-down assay between GST and histone H3 or H3K4me3 was performed. The bottom panel of each figure is a loading control. Unprocessed blots were provided in Supplementary Fig. 4.
Histone H3K4me3 status in WRKY70
WRKY70 is a transcription factor that positively regulates SA-responsive genes, and its expression is promoted by increased levels of salicylic acid38. The siz1-2 mutant accumulates salicylic acid and exhibits a dwarf-like phenotype7,39. Thus, histone H3K4me3 status in the WRKY70 gene was investigated by chromatin immunoprecipitation (ChIP) assay. Under normal conditions, histone H3K4me3 did not accumulate in WRKY70 in the wild type (Fig. 7a). Under cold treatment, histone H3K4me3 was detected in the wild type (Fig. 7a). This is because salicylic acid accumulation was promoted by cold stress, as described previously40. On the other hand, the histone H3K4me3 was highly detected in the siz1-2 mutant under both normal and low-temperature conditions (Fig. 7a). The expression level of WRKY70 was investigated. The transcription of WRKY70 was slightly induced by cold stress (Fig. 7b). WRKY70 expression was higher in the siz1-2 mutant before and after cold treatment and low expression was observed in the atx1 mutant (Fig. 7b). The expression level of WRKY70 was correlated with the status of histone H3K4me3. These results suggest that high levels of histone H3K4me3 in the siz1-2 mutant enhanced expression of the WRKY70 gene.
Fig. 7. H3K4me3 levels and transcription of WRKY70 in the siz1-2 mutant.
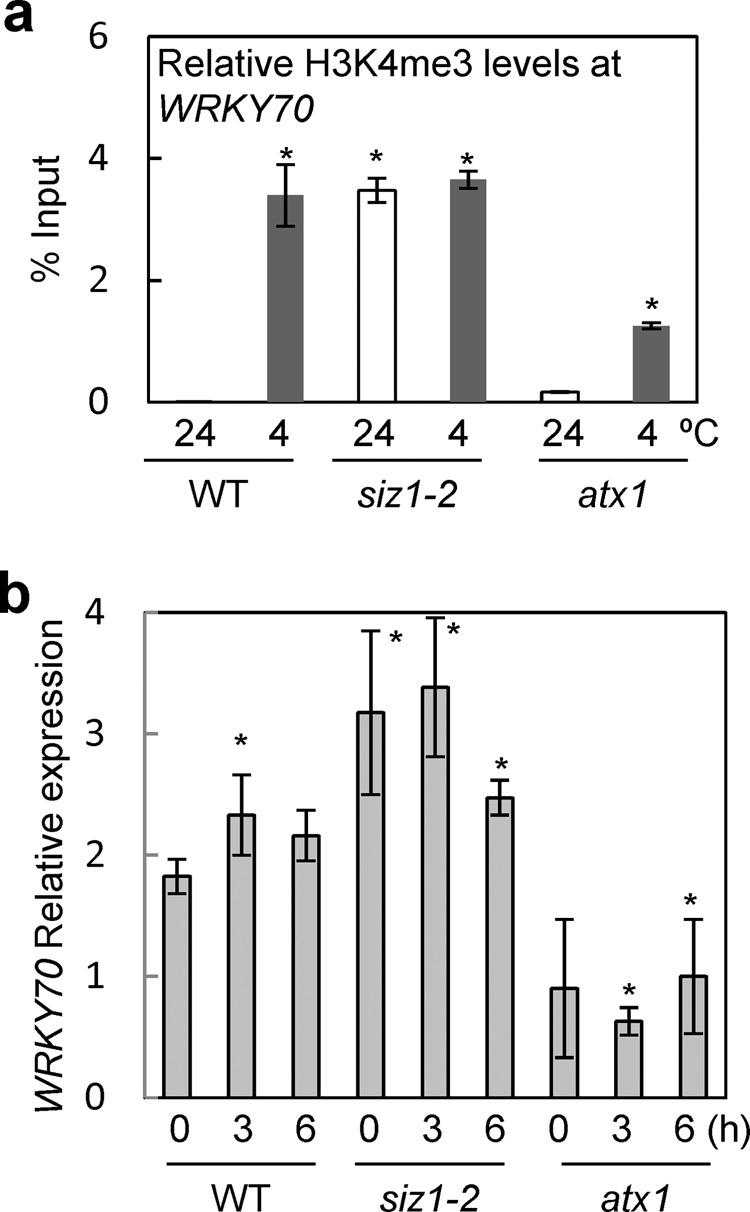
a Levels of histone H3K4me3 at the WRKY70 gene in wild type and the siz1-2 mutant. Wild type, the siz1-2 mutant, and the atx1 mutant were grown for 3 weeks at 24 °C. The plants were then subjected to cold treatment at 4 °C for 3 h. Then, a ChIP assay was performed. The atx1 mutant was used as a control to demonstrate that that levels of histone H3K4me3 were low compared with wild type, as described previously69. b WRKY70 expression was detected in wild type, the siz1-2 mutant, and the atx1 mutant with or without cold treatment. Each experiment was repeated three times and representative data are shown. Each bar represents the standard error of the mean (± SE, n = 3). Asterisks indicate a statistical difference from the wild type without cold treatment (p < 0.05) as determined by Student’s t test.
Interaction between SIZ1 and ATX1
As shown above, PHD preferentially recognized histone H3K4me3 (Fig. 6), and dysfunction of SIZ1 enhanced H3K4me3 status in the WRKY70 promoter (Fig. 7). SIZ1 may inhibit methylation of histone H3K4 and prevent transcription initiation via interaction of histone H3K4me3, which is a prominent histone mark associated with transcriptionally active genes41. Among histone lysine methyltransferases, ATX1 mediates H3K4 trimethylation and ATX2 mediates H3K4 dimethylation34,35. Concurrent disruption of the ATX3, ATX4, and ATX5 genes significantly reduced H3K4me2 and H3K4me3 levels, suggesting that these are redundant for H3K4 di- and trimethylation42. These previous results suggest that ATX1–5 are involved in the transfer of a methyl group to histone H3K4. Thus, we assumed that the PHD finger of SIZ1 interacts with ATX to block activity of histone lysine methyltransferase. ATX1–5 proteins contain catalytic SET domains, which bind S-adenosylmethionine and the substrate lysine43. The SET domains of ATX proteins fused with maltose-binding protein (MBP) were purified, and each MBP-ATXSET was incubated with GST-PHD. After pull-down with GST-PHD, MBP-ATXSET was detected with an anti-MBP antibody. All SET domains were detected by western blot analysis (Fig. 8), indicating that the PHD of SIZ1 was able to interact with the SET domain of each ATX protein. Because GST-PHD(C162S) also detected all SET domains, substitution of C162 in the PHD domain did not affect binding activity to the SET domain of each ATX protein. Probably, PHD has different binding activity to histone H3K4me3 or ATX proteins.
Fig. 8. The PHD finger of SIZ1 interacts with the SET domain of ATX proteins.
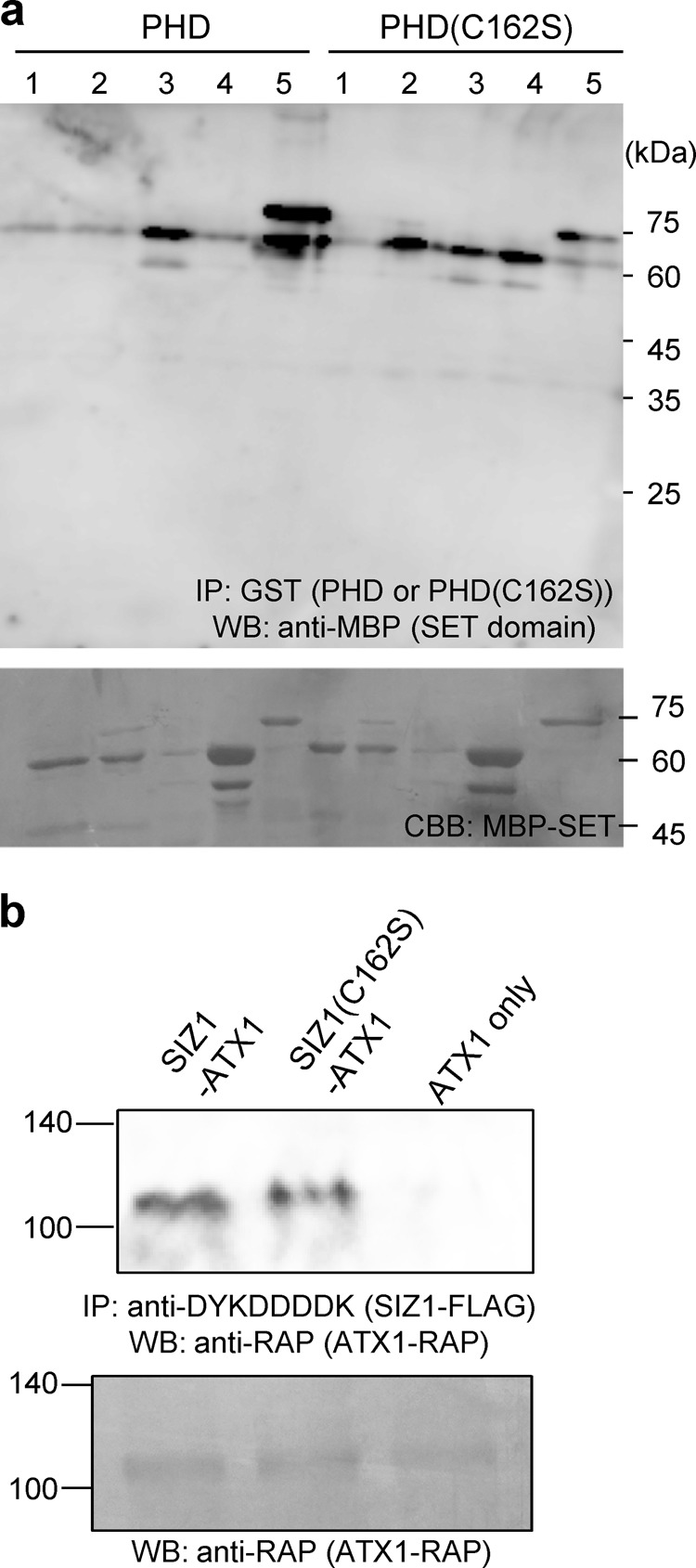
a GST-PHD or GST-PHD(C162S) and MBP-ATXSET were incubated and pull-down of GST-PHD or GST-PHD(C162S) was performed with glutathione Sepharose beads. Then, MBP-ATXSET proteins were detected by immunoblot analysis with an anti-MBP monoclonal antibody. Before immunoprecipitation, 5% of protein mixture was loaded onto SDS–PAGE and the gel was stained with Coomassie Brilliant Blue to detect MBP-SET domain of each ATX protein. b Interaction between SIZ1 or SIZ1(C162S) and ATX1 in vivo. Entire coding sequence of SIZ1 and ATX1, which are fused with FLAG and RAP, respectively, was transiently expressed in N. benthamiana. The immunoprecipitation was performed with anti-DYKDDDDK antibody magnetic beads and the immunoprecipitant was detected by anti-RAP antibody. Unprocessed blot was provided in Supplementary Fig. 4.
To confirm interaction in vivo, full-length coding sequence of SIZ1 including the PHD finger or ATX1 including the SET domain was inserted into pBYR2HS44 fused with the FLAG tag or the RAP tag, respectively. SIZ1-FLAG or SIZ1(C162S)-FLAG was immunoprecipitated with anti-DYKDDDK antibody. The immunoprecipitant was detected with anti-RAP tag antibody45. These results suggest that SIZ1 interacts with ATX1 in vivo.
Discussion
SIZ1 is an important SUMO E3 ligase in plants, and only plant SIZ proteins contain a PHD finger46. In the present study, we demonstrated that this PHD finger is biologically important, because a mutation or deletion of the PHD finger from SIZ1 was unable to complement the siz1-2 mutation (Figs. 1–4). Furthermore, the PHD finger preferentially recognized trimethylated histone H3K4 (Fig. 6a, b). Substitution of C162 with S prevented interaction between the PHD finger and H3K4me3 (Fig. 6c), suggesting that inhibition of PHD (C162S)-binding activity may prevent complementation of the siz1-2 mutation. The PHD finger also interacted with ATX proteins (Fig. 8), which methylate histone H3K4, probably resulting in suppressed methylation function of ATX proteins.
Histone H3K4me3 is a landmark of active gene expression47. The recognition of H3K4me3 by PHD was first reported in human BPTF and ING226,27,33,48. PHD recognizes H3K4me3, but it does not affect transcriptional activation or repression, which is dependent on the recruitment of histone acetyl transferase or a histone deacetylase complex. Our results suggest that recognition of H3K4me3 by Arabidopsis SIZ1 induced transcriptional repression of WRKY70, because the siz1-2 mutation promoted its expression (Fig. 7). In mammalian cells, sumoylated histone H4, which is associated with gene repression, in nucleosomes activates LSD1 (lysine-specific demethylase1) by a mechanism dependent on the SUMO-interacting motif in CoREST (co-repressor for element 1 silencing transcription factor)49. LSD1 demethylates methylated histone H3K4 to downregulate gene expression. Sumoylated histone H4 enhances development of the LSD1–CoREST complex to repress gene activity. It is plausible that SIZ1-bound histone H3K4me3 may recruit demethylase or a co-repressor to repress gene expression in plants.
Although several reports have demonstrated that PHD fingers interact with H3K4me2/3, some bind to other histone marks, such as H3K9me350, H3K36me351, H3R252, or unmodified H3K453,54. PHD fingers are widely conserved in eukaryotic organisms, including plant species. PHD-containing proteins in plants are involved in the regulation of various biological functions, such as pathogen defense responses, developmental processes, and flowering time55. These results suggest that PHD has regulatory functions and is involved in the mediation of cross-talk between the epigenetic status of chromatin, and signaling and cellular pathways.
The PHD finger of Arabidopsis SIZ1 interacts with the SUMO E2 enzyme AtSCE1 and the bromodomain global transcription factor group E (GTE)30. The PHD contributes partially to sumoylation of AtSCE1 but is indispensable for sumoylation of GTE30. Similarly, the PHD finger of the human KRAB-associated protein 1 (KAP1) co-repressor binds to the SUMO E2 enzyme Ubc9 and functions as an intramolecular SUMO E3 ligase56. The PHD-mediated sumoylation of the adjacent bromodomain is required for gene silencing56,57. Conversely, the PHD finger of rice SIZ1 was unable to bind to SUMO E229. These results suggest that the tandem PHD finger-bromodomain, or association between the PHD finger with the bromodomain, is required for transcriptional silencing.
The bromodomain typically recognizes acetylated histones to interpret HAT activity and recognize the histone code58. The bromodomain is often found in combination with the PHD finger, as described above. And the bromodomain is also found in combination with other histone-binding domains, including the MBT and WD40 domains58. These domains recognize multiple modifications to regulate transcriptional activation or silencing. The NURF chromatin-remodeling complex subunit BPTF harbors a bromodomain and a PHD finger to interact with H4K16ac and H3K4me3, respectively, in the same nucleosome59. Conversely, lysine methylation is mediated by SET domain proteins, which are identified in Drosophila Su(var)3-9, Ez (Enhancer of Zesta), and Trithorax60. Su(var)3-9 methylates histone H3K9, whereas Ez methylates H3K2761. Trithorax is more likely to mediate methylation of histone H3K4. Based on similarity, five ATX (Arabidopsis trithorax) and two ATX-related proteins (ATXR3 and ATXR7) have been proposed as H3K4 methyltransferases62. In fact, ATX1, ATX2, and ATXR7 have been shown to be involved in FLC activation and histone methylation35,63. To suppress transcription, histone methylation at histone H3K4 should be blocked. The SET domains of ATX proteins interacted with the PHD finger of SIZ1 (Fig. 8). It is likely that the PHD finger of SIZ1 interacts with ATX proteins to inhibit their access to histone H3K4.
The PHD finger of SIZ1 recognizes histone H3K4me3 and this interaction may suppress transcriptional activation. The PHD finger also binds to ATXs and this interaction may repress methyltransferase activity for further activation of transcription. Because PHD(C162S) was unable to complement the siz1-2 mutation (Figs. 2–4) and was unable to interact with histone H3K4me3 (Fig. 6) but bound to ATXs (Fig. 8), recognition of histone H3K4me3 is more important for regulation of transcription.
In conclusion, the PHD finger of SIZ1 is important for recognition of the histone code, H3K4me3, to suppress transcription of WRKY70 and for interaction with ATX proteins, probably to inhibit their binding to histone H3K4.
Methods
Plant materials and physiological analysis
The Arabidopsis T-DNA insertion mutants siz1-215 and siz1-2 were transformed with pCambia1302-AtSIZ1full::AtSIZ1:GFP and the resulting plants, siz1-2 with ProSIZ1::SIZ1:GFP31 were on the Col-0 background.
To produce siz1-2 with ProSIZ1::SIZ1(ΔPHD):GFP, the 5′- or 3′-regions of SIZ1 were amplified with the primers, SIZ1-2hyF and SIZ1-PHDdeltaR, or SIZ1-PHDdeltaF and SIZ1-2hyR (Supplementary Table 1), respectively. After purification of PCR products, SIZ1(ΔPHD) was amplified with the primers, SIZ1-2hyF and SIZ1-2hyR, with the 5′- and 3′-regions of SIZ1 as templates. The PCR product was digested with SalI and PstI and the resulting product was inserted with SalI- and PstI-digested pCambia1302-AtSIZ1full::AtSIZ1:GFP. The resulting plasmid, pCambia1302-AtSIZ1full::AtSIZ1(ΔPHD):GFP, was transformed into Agrobacterium and then transgenic Col-0 plants containing ProSIZ1::SIZ1(ΔPHD):GFP were produced. The transgenic Col-0 plant was crossed with the siz1-2 mutant. In the F2 population, plants harboring both homozygotes of the siz1-2 mutation and ProSIZ1::SIZ1(ΔPHD):GFP were selected and named siz1-2 with ProSIZ1::SIZ1(ΔPHD):GFP. The siz1-2 mutant or wild-type SIZ1 was determined with the primers, LP034008 and Salk_Lba1 or LP034008 and RP0034008 (Supplementary Table 1), respectively. Similarly, pCambia1302-AtSIZ1full::AtSIZ1(C162S):GFP or pCambia1302-AtSIZ1full::AtSIZ1(C117S):GFP was produced with the primers, SIZ1-2hyF, AtSIZ1-C162S-R, AtSIZ1-C162S-F, and SIZ1-2hyR, or SIZ1-2hyF, AtSIZ1-C117S-R, AtSIZ1-C117S-F, and SIZ1-2hyR (Supplementary Table 1), respectively. These vectors were transformed and siz1-2 with ProSIZ1::SIZ1(C162S):GFP or siz1-2 with ProSIZ1::SIZ1(C117S):GFP were produced. Arabidopsis plants were grown in soil or Petri dishes at 24 °C under a long-day photoperiod (16 h light/8 h dark).
To examine the effect of ABA hypersensitivity on root growth, the seeds were surface-sterilized and sown onto half Murashige and Skoog (MS) medium containing 1% sucrose and 0.8% agar. Three-day-old seedlings were transferred onto media supplemented with 0, 5, 10, or 15 µm ABA (Sigma). Root growth was measured as the difference in root length between the beginning and end of the growth evaluation period24.
To evaluate freezing sensitivity, the plants were grown at 24 °C for 3 weeks in soil and, then incubated at 4 °C for 1 week for acclimation to low temperature. After acclimation, plants were incubated at 0 °C for 1 h, and the temperature was lowered by 2 °C h−1 until it reached to − 6 °C and was maintained for 4 h in the incubator (IN602, Yamato Scientific Co., Ltd., Tokyo, Japan). Then, plants were incubated at 4 °C overnight and transferred to 24 °C. The survival ratio was determined 10 days after the freezing test was performed, as described previously64.
For the drought-tolerance test, water was withheld from 2-week-old plants for 2 weeks. Then, watering was resumed. To quantitatively determine the survival ratio, 16 plants of each genotype were grown in the same pot. After water was withheld followed by re-watering, the survival ratio was calculated as described previously4.
These physiological analyses were performed three or more times and representative data were shown in figures.
RNA preparation and RT-PCR
Total RNA was isolated using TRIzol reagent (Thermo Fisher Scientific), according to the manufacturer’s protocol. RT-PCR was performed as described previously65. The primers LP023805 and RP023805 (Supplementary Table 2) were used to detect SIZ1 expression. The primers as described in the previous work66 were used for detection of WRKY70 expression. Data are a representative result from three independent experiments.
Pull-down analysis
The PHD region was amplified with the primers pGEX5X-PHD-F and pGEX5X-PHD-R (Supplementary Table 1). The PCR product was inserted into EcoRI- and SalI-digested pGEX5X-1 (GE healthcare) via the In-Fusion reaction (Takara Bio). The vector was transformed into Escherichia coli Rosetta-Gami(DE3)pLysS (Novagen), and GST-PHD protein was purified with Glutathione Sepharose 4 Fast Flow (GE healthcare) according to the manufacturer’s protocol. One-microgram of GST-PHD was incubated with 0.5 µg of biotinylated histone H3 variant (Active Motif) in 300 µL of binding buffer (50 mm Tris-HCl, pH 7.8, 300 mm NaCl, 0.1% NP-40, and 1 mm PMSF) overnight. Biotinylated histone H3 variant was pulled down with streptavidin beads. The sample was separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE), and western blot analysis was performed with anti-GST antibody. For input, 10 ng of GST-PHD was loaded. For loading control, 0.5% of mixture before pull-down was loaded onto SDS–PAGE and GST-PHD was detected by anti-GST antibody.
The SET domains of ATX proteins were amplified with the primers, ATX(1 or 2)SET-F-SalI and ATX(1 or 2)-SalI-R (Supplementary Table 1), respectively. The SalI-digested PCR products were inserted into the SalI-digested pMAL-c2X (New England Biolabs) to produce pMAL-ATX1SET and pMAL-ATX2SET. The SET domains of ATX3–5 were amplified with the primers, ATX(3–5)-F-BamHI and ATX(3–5)-BamHI-R (Supplementary Table 1), respectively. The PCR products and pMAL-c2X were digested with BamHI and ligated. The resulting vectors were named pMAL-ATX(3–5)SET. The vectors were transformed into E. coli Rosetta-Gami(DE3)pLysS (Novagen), and MBP-ATXSET protein was purified with amylose resin (New England Biolabs) according to the manufacturer’s protocol. One-microgram of GST-PHD and 1 µg of MAL-ATXSET were incubated in 300 µL of peptide-binding buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.1% NP-40) overnight. GST-PHD was pulled down with glutathione Sepharose beads. The sample was separated with SDS–PAGE and western blot analysis was performed with an anti-MBP monoclonal antibody (New England Biolabs).
For all immunoblot analysis, the primary antibody was used with 20,000 dilution and the secondary antibody was used with 10,000 dilution.
Co-immunoprecipitation assay
The FLAG tag or the RAP tag was introduced into the high-level transient expression ‘Tsukuba system’44,67. The PCR products containing His-tag (HHHHHH), and FLAG tag (DYKDDDDK) were produced with the primers pBYR2HS-Flag-F, FLAG-His, and pBYR2HS-FlagHis-R (Supplementary Table 1). The PCR products containing His-tag (HHHHHH), RAP tag (DMVNPGLEDRIE)45, and the recognition site for HRV 3 C protease (LEVLFQGP), were produced with the primers pBYR2HS-HRV3C-F, HRV3C-RAP-His, and pBYR2HS-stopHis-R (Supplementary Table 1). These PCR products were introduced into the SalI-digested pBYR2HS with an In-Fusion HD Cloning Kit. The resulting constructs were designated as pBYR2HS-CFH or pBYR2HS-CRH, respectively. The coding sequence of SIZ1 or ATX1 was amplified with the primers, pBYR2HS-AtSIZ1-F and pBYR2HS-AtSIZ1-R, or pBYR2HS-ATX1-F and pBYR2HS-ATX1-R (Supplementary Table 1), respectively. The resulting PCR products were introduced into the SalI-digested pBYR2HS-CFH or pBYR2HS-CRH. pBYR2HS-SIZ1-FH, pBYR2HS-SIZ1(C162S)-FH, or pBYR2HS-ATX1-RH was transformed into Agrobacterium tumefaciens GV3101, then, agroinfiltration was performed to Nicotiana benthamiana44. Soluble protein solution was prepared with lysis buffer65. SIZ1-FH or SIZ1(C162S)-FH was immunoprecipitated with anti-DYKDDDDK tag antibody magnetic beads (Fujifilm Wako Pure Chemical Industries). The immunoprecipitant was separated by SDS–PAGE and immunoblot analysis with an anti-RAP tag antibody (PMab2)45.
Chromatin immunoprecipitation (ChIP) assay
To investigate histone H3K4me3 status in the promoter of WRKY70, a ChIP assay was performed as described previously, with modification68. Three-week-old wild-type siz1-2, and atx1 plants (SALK_149002C) were treated with or without cold (4 °C) for 3 h, and the seedlings were subsequently harvested and cross-linked with buffer A (0.4 m sucrose; 10 mm Tris-HCl, pH 8.0; 1 mm EDTA; 1 mm PMSF; 1% formaldehyde). The cross-linking reaction was stopped with 125 mm glycine. The tissues were ground in liquid nitrogen, resuspended in lysis buffer (50 mm HEPES, pH 7.5; 150 mm NaCl; 1 mm EDTA; 1% Triton X-100; 0.1% deoxycholate; 0.1% SDS; 1 mm PMSF; and 1 × protease inhibitor cocktail [Nacalai Tesque]), and sonicated (Microson XL-2000 sonicator, Qsonica, LLC., USA) to achieve an average fragment size of 0.1–1.0 kb. After sonication, centrifugation was performed, and the supernatants were incubated overnight at 4 °C with anti-histone H3K4me3 monoclonal antibody (Active Motif). Immunoprecipitation was performed using Magna ChIP A Chromatin Immunoprecipitation Kits (Millipore). Quantitative PCR was performed with THUNDERBIRD SYBR Premix (Toyobo) using a real-time PCR Thermal Cycler Dice (Takara Bio). The primers, WRKY70-ChipF and WRKY70-ChipR (Supplementary Table 2), were used for detection. The relative quantities of immunoprecipitated DNA fragments were calculated as the percentage of input chromatin that was immunoprecipitated using the comparative CT method. Data are a representative experiment from three independent experiments.
Statistics and reproducibilty
Statistical analysis was performed using Excel software (Microsoft) or IBM SPSS Statics. P values < 0.05 were considered significant.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We thank Ms. Kazuko Ito, Ms. Rieko Nozawa, and Ms. Yuri Nemoto at Tsukuba-Plant Innovation Research Center (T-PIRC), University of Tsukuba for technical support. This work was supported by JSPS Grant-in-Aid for Scientific Research on Innovative Areas (JP16H01458 and JP19H04637) and by a Cooperative Research Grant from the Plant Transgenic Design Initiative, Gene Research Center, T-PIRC, University of Tsukuba.
Author contributions
K.M. designed the study. N.R. and K.M performed most of experiments. T.S. and K.M. contributed reagents and materials and analyzed data. K.M. wrote the manuscript.
Data availability
The authors declare that all data supporting the findings of this study are available within the paper and its supplementary information files.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Kenji Miura, Na Renhu.
Supplementary information
Supplementary information is available for this paper at 10.1038/s42003-019-0746-2.
References
- 1.Hendriks IA, Vertegaal AC. A comprehensive compilation of SUMO proteomics. Nat. Rev. Mol. Cell Biol. 2016;17:581–595. doi: 10.1038/nrm.2016.81. [DOI] [PubMed] [Google Scholar]
- 2.Augustine RC, Vierstra RD. SUMOylation: re-wiring the plant nucleus during stress and development. Curr. Opin. Plant Biol. 2018;45:143–154. doi: 10.1016/j.pbi.2018.06.006. [DOI] [PubMed] [Google Scholar]
- 3.Zhang S, et al. A novel tomato SUMO E3 ligase, SlSIZ1, confers drought tolerance in transgenic tobacco. J. Integr. Plant Biol. 2017;59:102–117. doi: 10.1111/jipb.12514. [DOI] [PubMed] [Google Scholar]
- 4.Miura K, et al. SIZ1 deficiency causes reduced stomatal aperture and enhanced drought tolerance via controlling salicylic acid-induced accumulation of reactive oxygen species in Arabidopsis. Plant J. 2013;49:79–90. doi: 10.1111/tpj.12014. [DOI] [PubMed] [Google Scholar]
- 5.Miura K, Sato A, Ohta M, Furukawa J. Increased tolerance to salt stress in the phosphate-accumulating Arabidopsis mutants siz1 and pho2. Planta. 2011;234:1191–1199. doi: 10.1007/s00425-011-1476-y. [DOI] [PubMed] [Google Scholar]
- 6.Miura K, et al. SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell. 2007;19:1403–1414. doi: 10.1105/tpc.106.048397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee J, et al. Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J. 2007;49:79–90. doi: 10.1111/j.1365-313X.2006.02947.x. [DOI] [PubMed] [Google Scholar]
- 8.Catala R, et al. The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses. Plant Cell. 2007;19:2952–2966. doi: 10.1105/tpc.106.049981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saleh A, et al. Posttranslational modifications of the master transcriptional regulator NPR1 enable dynamic but tight control of plant immune responses. Cell Host Microbe. 2015;18:169–182. doi: 10.1016/j.chom.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng Y, Schumaker KS, Guo Y. Sumoylation of transcription factor MYB30 by the small ubiquitin-like modifier E3 ligase SIZ1 mediates abscisic acid response in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 2012;109:12822–12827. doi: 10.1073/pnas.1202630109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van den Burg HA, Kini RK, Schuurink RC, Takken FL. Arabidopsis small ubiquitin-like modifier paralogs have distinct functions in development and defense. Plant Cell. 2010;22:1998–2016. doi: 10.1105/tpc.109.070961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miura K, et al. Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc. Natl. Acad. Sci. USA. 2009;106:5418–5423. doi: 10.1073/pnas.0811088106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colby T, Matthai A, Boeckelmann A, Stuible HP. SUMO-conjugating and SUMO-deconjugating enzymes from Arabidopsis. Plant Physiol. 2006;142:318–332. doi: 10.1104/pp.106.085415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saracco SA, Miller MJ, Kurepa J, Vierstra RD. Genetic analysis of SUMOylation in Arabidopsis: conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol. 2007;145:119–134. doi: 10.1104/pp.107.102285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miura K, et al. The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc. Natl. Acad. Sci. USA. 2005;102:7760–7765. doi: 10.1073/pnas.0500778102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishida T, et al. SUMO E3 Ligase HIGH PLOIDY2 regulates endocycle onset and meristem maintenance in Arabidopsis. Plant Cell. 2009;21:2284–2297. doi: 10.1105/tpc.109.068072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hay RT. Decoding the SUMO signal. Biochem. Soc. Trans. 2013;41:463–473. doi: 10.1042/BST20130015. [DOI] [PubMed] [Google Scholar]
- 18.Miller MJ, Barrett-Wilt GA, Hua Z, Vierstra RD. Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2010;107:16512–16517. doi: 10.1073/pnas.1004181107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rytz TC, et al. SUMOylome profiling reveals a diverse array of nuclear targets modified by the SUMO Ligase SIZ1 during heat stress. Plant Cell. 2018;30:1077–1099. doi: 10.1105/tpc.17.00993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang L, et al. The Arabidopsis SUMO E3 ligase AtMMS21, a homologue of NSE2/MMS21, regulates cell proliferation in the root. Plant J. 2009;60:666–678. doi: 10.1111/j.1365-313X.2009.03992.x. [DOI] [PubMed] [Google Scholar]
- 21.Tomanov K, et al. Arabidopsis PIAL1 and 2 promote SUMO chain formation as E4-type SUMO ligases and are involved in stress responses and sulfur metabolism. Plant Cell. 2014;26:4547–4560. doi: 10.1105/tpc.114.131300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishida T, Yoshimura M, Miura K, Sugimoto K. MMS21/HPY2 and SIZ1, two Arabidopsis SUMO E3 ligases, have distinct functions in development. PloS ONE. 2012;7:e46897. doi: 10.1371/journal.pone.0046897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomanov K, et al. Arabidopsis PIAL1 and 2 promote SUMO chain formation as E4-type SUMO ligases and are involved in stress responses and sulfur metabolism. Plant Cell. 2014;26:4547–4560. doi: 10.1105/tpc.114.131300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miura K, Nozawa R. Overexpression of SIZ1 enhances tolerance to cold and salt stresses and attenuates response to abscisic acid in Arabidopsis thaliana. Plant. Biotechnol. 2014;31:167–172. doi: 10.5511/plantbiotechnology.14.0109a. [DOI] [Google Scholar]
- 25.Cheong MS, et al. Specific domain structures control abscisic acid-, salicylic acid-, and stress-mediated SIZ1 phenotypes. Plant Physiol. 2009;151:1930–1942. doi: 10.1104/pp.109.143719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pena PV, et al. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006;442:100–103. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, et al. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y, Li H. Many keys to push: diversifying the ‘readership' of plant homeodomain fingers. Acta Biochim. Biophys. Sin. 2012;44:28–39. doi: 10.1093/abbs/gmr117. [DOI] [PubMed] [Google Scholar]
- 29.Shindo H, et al. PHD finger of the SUMO ligase Siz/PIAS family in rice reveals specific binding for methylated histone H3 at lysine 4 and arginine 2. FEBS Lett. 2012;586:1783–1789. doi: 10.1016/j.febslet.2012.04.063. [DOI] [PubMed] [Google Scholar]
- 30.Garcia-Dominguez M, March-Diaz R, Reyes JC. The PHD domain of plant PIAS proteins mediates sumoylation of bromodomain GTE proteins. J. Biol. Chem. 2008;283:21469–21477. doi: 10.1074/jbc.M708176200. [DOI] [PubMed] [Google Scholar]
- 31.Jin JB, et al. The SUMO E3 ligase, AtSIZ1, regulates flowering by controlling a salicylic acid-mediated floral promotion pathway and through affects on FLC chromatin structure. Plant J. 2008;53:530–540. doi: 10.1111/j.1365-313X.2007.03359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang G, et al. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc. Natl. Acad. Sci. USA. 2004;101:7357–7362. doi: 10.1073/pnas.0401866101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi X, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saleh A, et al. The highly similar Arabidopsis homologs of trithorax ATX1 and ATX2 encode proteins with divergent biochemical functions. Plant Cell. 2008;20:568–579. doi: 10.1105/tpc.107.056614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pien S, et al. ARABIDOPSIS TRITHORAX1 dynamically regulates FLOWERING LOCUS C activation via histone 3 lysine 4 trimethylation. Plant Cell. 2008;20:580–588. doi: 10.1105/tpc.108.058172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pascual J, Martinez-Yamout M, Dyson HJ, Wright PE. Structure of the PHD zinc finger from human Williams-Beuren syndrome transcription factor. J. Mol. Biol. 2000;304:723–729. doi: 10.1006/jmbi.2000.4308. [DOI] [PubMed] [Google Scholar]
- 37.Miura K, Hasegawa PM. Sumoylation and abscisic acid signaling. Plant Signal Behav. 2009;4:1176–1178. doi: 10.4161/psb.4.12.10044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J, Brader G, Palva ET. The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell. 2004;16:319–331. doi: 10.1105/tpc.016980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miura K, Lee J, Miura T, Hasegawa PM. SIZ1 controls cell growth and plant development in Arabidopsis through salicylic acid. Plant Cell Physiol. 2010;51:103–113. doi: 10.1093/pcp/pcp171. [DOI] [PubMed] [Google Scholar]
- 40.Scott IM, Clarke SM, Wood JE, Mur LA. Salicylate accumulation inhibits growth at chilling temperature in Arabidopsis. Plant Physiol. 2004;135:1040–1049. doi: 10.1104/pp.104.041293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol. Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 42.Chen LQ, et al. ATX3, ATX4, and ATX5 encode putative H3K4 methyltransferases and are critical for plant development. Plant Physiol. 2017;174:1795–1806. doi: 10.1104/pp.16.01944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trievel RC, Beach BM, Dirk LM, Houtz RL, Hurley JH. Structure and catalytic mechanism of a SET domain protein methyltransferase. Cell. 2002;111:91–103. doi: 10.1016/S0092-8674(02)01000-0. [DOI] [PubMed] [Google Scholar]
- 44.Yamamoto T, et al. Improvement of the transient expression system for production of recombinant proteins in plants. Sci. Rep. 2018;8:4755. doi: 10.1038/s41598-018-23024-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fujii Y, et al. Development of RAP tag, a novel tagging system for protein detection and purification. Monoclon. Antib. Immunodiagn. Immunother. 2017;36:68–71. doi: 10.1089/mab.2016.0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miura K, Hasegawa PM. Sumoylation and other ubiquitin-like post-translational modifications in plants. Trends Cell Biol. 2010;20:223–232. doi: 10.1016/j.tcb.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 47.Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wysocka J, et al. PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 49.Dhall A, Weller CE, Chu A, Shelton PMM, Chatterjee C. Chemically sumoylated histone H4 stimulates intranucleosomal demethylation by the LSD1–CoREST complex. ACS Chem. Biol. 2017;12:2275–2280. doi: 10.1021/acschembio.7b00716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arita K, et al. Recognition of modification status on a histone H3 tail by linked histone reader modules of the epigenetic regulator UHRF1. Proc. Natl. Acad. Sci. USA. 2012;109:12950–12955. doi: 10.1073/pnas.1203701109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi X, et al. Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J. Biol. Chem. 2007;282:2450–2455. doi: 10.1074/jbc.C600286200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chignola F, et al. The solution structure of the first PHD finger of autoimmune regulator in complex with non-modified histone H3 tail reveals the antagonistic role of H3R2 methylation. Nucleic Acids Res. 2009;37:2951–2961. doi: 10.1093/nar/gkp166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ooi SK, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsai WW, et al. TRIM24 links a non-canonical histone signature to breast cancer. Nature. 2010;468:927–932. doi: 10.1038/nature09542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mouriz A, López-González L, Jarillo JA, Piñeiro M. PHDs govern plant development. Plant Signal Behav. 2015;10:e993253–e993253. doi: 10.4161/15592324.2014.993253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ivanov AV, et al. PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol. Cell. 2007;28:823–837. doi: 10.1016/j.molcel.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zeng L, et al. Structural insights into human KAP1 PHD finger-bromodomain and its role in gene silencing. Nat. Struct. Mol. Biol. 2008;15:626–633. doi: 10.1038/nsmb.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Josling GA, Selvarajah SA, Petter M, Duffy MF. The role of bromodomain proteins in regulating gene expression. Genes. 2012;3:320–343. doi: 10.3390/genes3020320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ruthenburg AJ, et al. Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell. 2011;145:692–706. doi: 10.1016/j.cell.2011.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bottomley MJ. Structures of protein domains that create or recognize histone modifications. EMBO Rep. 2004;5:464–469. doi: 10.1038/sj.embor.7400146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rea S, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 62.Ng DWK, et al. Plant SET domain-containing proteins: structure, function and regulation. Biochim. Biophys. Acta. 2007;1769:316–329. doi: 10.1016/j.bbaexp.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tamada Y, Yun JY, Woo SC, Amasino RM. Arabidopsis trithorax-related7 is required for methylation of lysine 4 of histone H3 and for transcriptional activation of flowering locus C. Plant Cell. 2009;21:3257–3269. doi: 10.1105/tpc.109.070060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mori K, et al. Ca2+-permeable mechanosensitive channels MCA1 and MCA2 mediate cold-induced cytosolic Ca2+ increase and cold tolerance in Arabidopsis. Sci. Rep. 2018;8:550. doi: 10.1038/s41598-017-17483-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miura K, et al. SlICE1 encoding a MYC-type transcription factor controls cold tolerance in tomato, Solanum lycopersicum. Plant Biotechnol. 2012;29:253–260. doi: 10.5511/plantbiotechnology.12.0303a. [DOI] [Google Scholar]
- 66.Chen J, et al. Arabidopsis WRKY46, WRKY54, and WRKY70 transcription factors are involved in brassinosteroid-regulated plant growth and drought responses. Plant Cell. 2017;29:1425–1439. doi: 10.1105/tpc.17.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Suzaki T, Tsuda M, Ezura H, Day B, Miura K. Agroinfiltration-based efficient transient protein expression in leguminous plants. Plant Biotechnol. 2019;36:119–123. doi: 10.5511/plantbiotechnology.19.0220b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ohta M, et al. MYC-type transcription factors, MYC67 and MYC70, interact with ICE1 and negatively regulate cold tolerance in Arabidopsis. Sci. Rep. 2018;8:11622. doi: 10.1038/s41598-018-29722-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ding Y, et al. ATX1-generated H3K4me3 is required for efficient elongation of transcription, not initiation, at ATX1-regulated genes. PLoS Genet. 2012;8:e1003111. doi: 10.1371/journal.pgen.1003111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the paper and its supplementary information files.