Fig. 1.
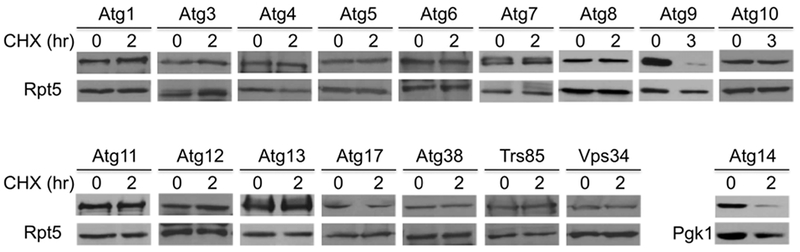
Two autophagy regulators are degraded under normal growth conditions. The protein stability of 17 autophagy regulators was examined in wild-type yeast cells by a protein expression shut-off assay. Yeast cells bearing GFP-Atg8 or MORF tagged autophagy proteins were grown to log phase, and samples were collected at indicated time points after cycloheximide (CHX) turned off protein synthesis. Extracts were analyzed by western blotting with anti-GFP for Atg8 or an HA antibody to detect MORF-tagged proteins. Equal loading was ascertained by immunoblotting with Rpt5 antibody (lower panels). For Atg14, we used Pgk1 as a loading control as the sizes of Atg14 and Rpt5 are similar. The identities of autophagy regulators are shown at the top.
