Fig. 2.
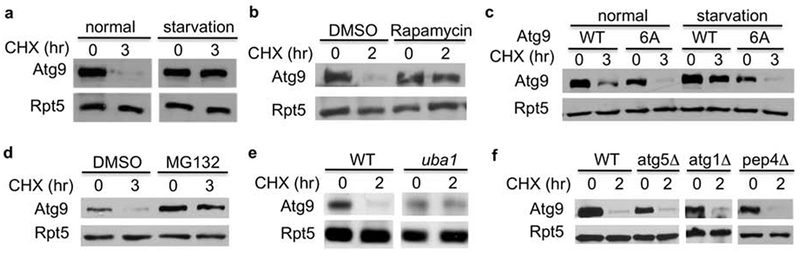
Atg9 turnover is hindered under cellular stress conditions that activate autophagy and regulated by the proteasome. (a,b) Immunoblot analysis of MORF-tagged Atg9 degradation in the absence or presence of rapamycin or nutrient depletion. (c) The degradation of Atg9 mutant defective for Atg1-phosphorylation is insensitive to starvation. The Atg9 mutant lacking Atg1 phosphorylation sites was previously dissected and the chromosomal copy of ATG9 is appended with a TAP tag at its C-terminus. The degradation of TAP-tagged wild-type and mutant Atg9 under the regulation of its endogenous promoter was analyzed with or without nutrient starvation. (d) Atg9 stability was evaluated in the presence and absence of the proteasome inhibitor MG132. (e) Atg9 degradation was assessed in wild-type and mutant cells bearing a temperature sensitive UBA1 mutation. The experiments were done as described above, except that cells were shifted to 37°C for I hr before the addition of cycloheximide. (f) Atg9 degradation was assessed in yeast cells lacking key autophagy regulators (i.e., ATG1, ATG5) or Pep4 protease.
