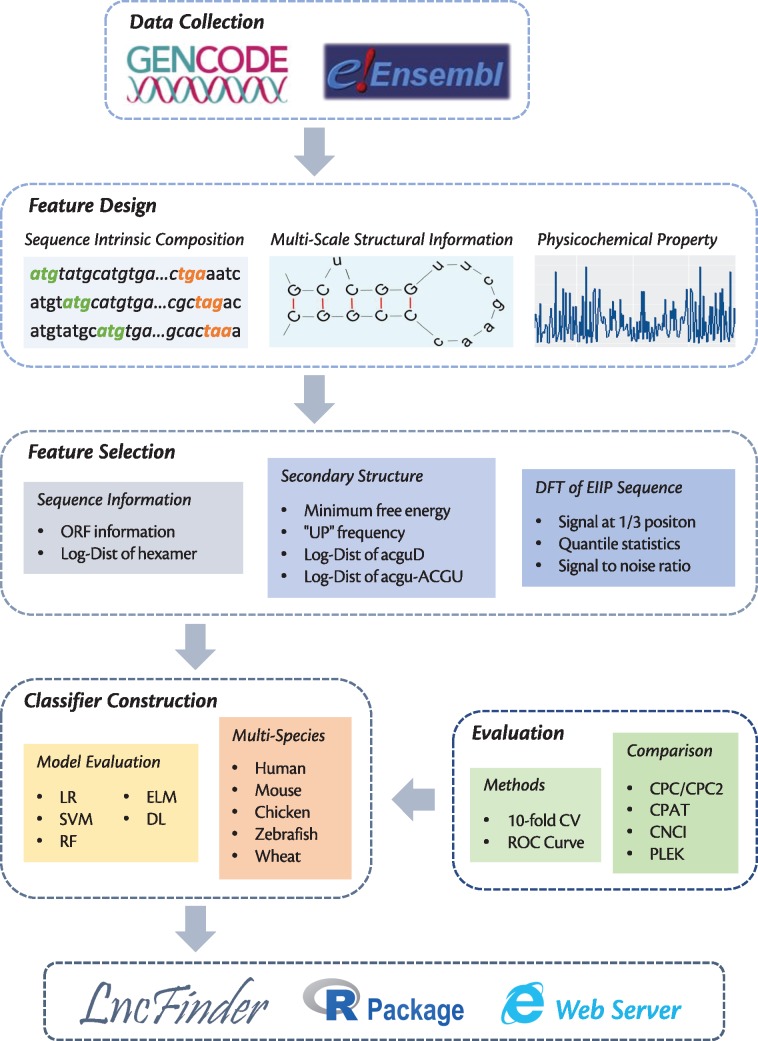
Figure 1.
Framework of this study. Data sets used in our experiments are collected from GENCODE and Ensembl. Only one transcript from each gene is used. In addition to sequence-intrinsic composition, features are also extracted from multi-scale secondary structure and EIIP-based physicochemical property using two feature selection schemes. Evaluated with 10-fold CV and ROC curve, the optimal feature combination and machine learning algorithm are obtained to develop a new method for lncRNA identification. This method is benchmarked against five popular tools on five species, and it is finally included in LncFinder, which is a highly flexible package for lncRNA identification and analysis. LncFinder is published as R package as well as web server.