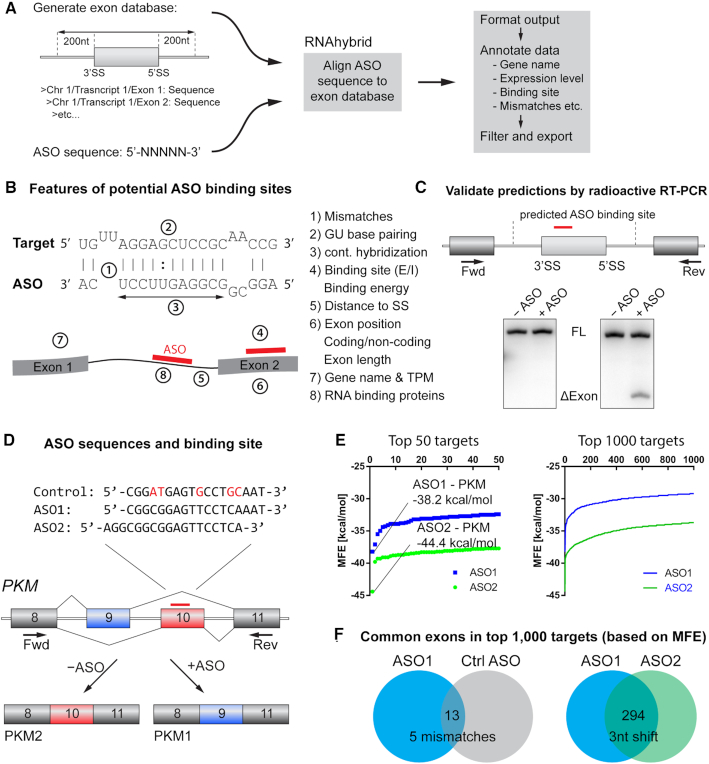
Figure 1.
Prediction of off-targets and validation by RT-PCR. (A) ASO sequences were aligned to an exon database, including 200 nt of each flanking intron. Sequence alignment was performed using RNAhybrid, allowing G:U wobble base pairing, internal loops, and bulges. The data were annotated and formatted for further analysis. (B) Features used to characterize the interaction between the ASO and the RNA target. (C) Potential off-targets were tested by radioactive RT-PCR, using primer pairs located at least 1 exon upstream and downstream of the binding site, respectively. Positive hits from the initial screen were analyzed further. (D) ASOs used in the initial off-target screen included two PKM splice-switching ASOs shifted by three nucleotides (ASO1-MOE and ASO2-MOE), and a control ASO with five mismatches to the target (Ctrl ASO-MOE). Nucleotides highlighted in red indicate mismatch positions. (E) Distribution of the minimum free energy (MFE; kcal/mol) of the top 1000 and top 50 hits from the RNAhybrid alignment prediction. PKM was the top hit for both ASO1 (−38.2 kcal/mol) and ASO2 (−44.4 kcal/mol). (F) Venn diagrams of overlapping targets among the top 1000 hits. ASO1 and the Ctrl ASO have 13 hits in common. ASO1 and ASO2 have 294 targets in common.