Figure 1.
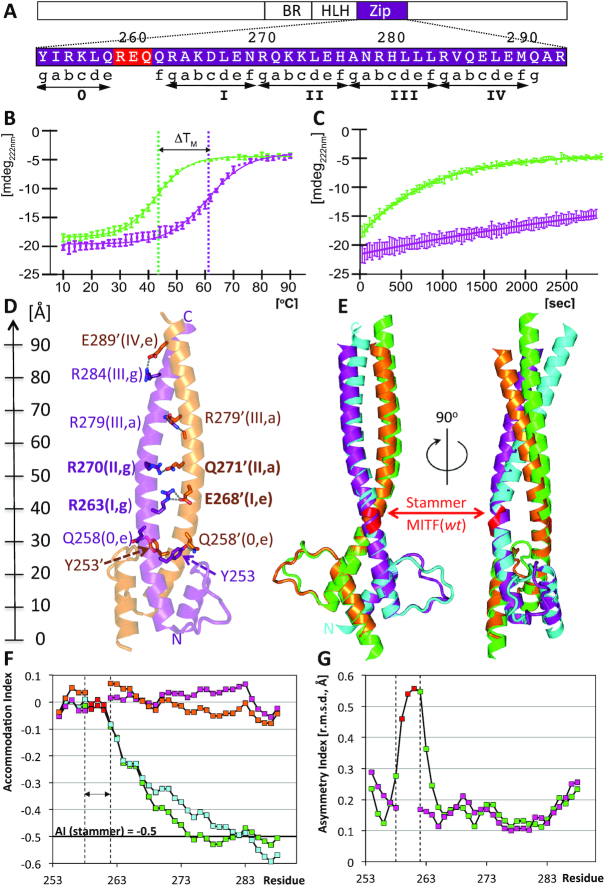
Structural and biophysical characterization of stammer-less MITF (Δ). (A) Zoom into the MITF sequence of the structurally characterized coiled coil region, indicating the relationship between residue positions, heptad numbers and positions. The stammer residues of MITF(wt) removed in MITF(Δ) are in red. Heptad repeats are indicated by ‘gabcdef’ positions and are numbered. The location of this region within the overall MITF domain structure is schematically shown above. (B) Thermal stability of MITF(wt, green) and MITF(Δ, purple) measured by circular dichroism. Thermal unfolding midpoints are indicated with vertical lines. The melting temperature of MITF(Δ) is increased by 18°C when compared to MITF(wt). (C) Differences in MITF(wt, green) and MITF(Δ, purple) susceptibility to proteolysis by proteinase K. (D) Ribbon representation showing specific coiled coil interactions. Residue numbers, heptad numbers and positions (cf. Figure 3) are indicated for all specific interactions observed. A ruler to the left indicates the dimensions of the MITF coiled coil. (E) Superposition of MITF(wt) (PDB code 4ATH) with stammer-less MITF(Δ) in two different orientations, rotated by 90 degrees around a vertical axis. Color codes: polypeptide chains A and B of MITF(Δ): purple, orange; polypeptide chains A and B of MITF(wt): green, cyan. Residues 259–261, which have been removed in the stammer-less MITF variant, are in red. The color codes are used also in subsequent figures. (F) Change of Accommodation Index (23) of the two polypeptide chains of each MITF(wt) and MITF(Δ) along the coiled coil segment. The three-residue segment removed in MITF(Δ) is indicated by vertical lines. The theoretical change in Accommodation Index of a stammer-containing coiled coil is −0,5, indicated by a horizontal line. (G) Change of Asymmetry Index measured by the change of root mean squares deviations using a residue window of 7 is plotted for MITF(wt) and MITF(Δ), indicating a peak asymmetry across the stammer segment.