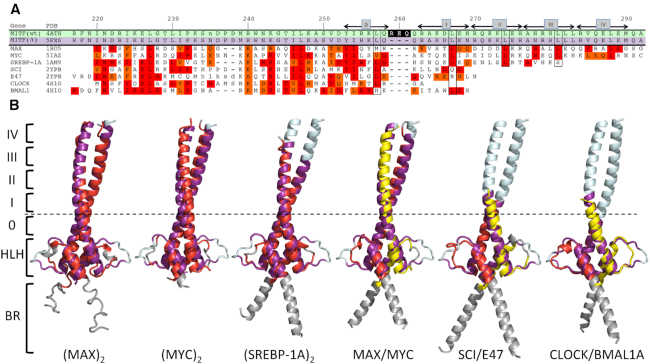
Figure 2.
Search for related bHLH-Zip transcription factors using MITF(Δ) as the template. (A) Structure-based sequence alignment. Gene names, as used in the text, and PDB codes of the target structures are indicated. Sequence numbers of the MITF sequence and the positions of heptad repeats 0, I–IV, as defined previously (22), are indicated above the alignment. Heptad positions d that are most conserved in known coiled coil structures are boxed to facilitate orientation. The three stammer residues that have been removed in MITF(Δ) are shown with a black background in the MITF(wt) sequence. Identical and conserved sequence positions are highlighted in red and orange, respectively. Note that the alignments for some bHLH-Zip transcription factors cover only a reduced number of heptad repeats. (B) Structural superimpositions. Color codes: MITF(Δ), purple (as in Figure 1) for superimposed parts, pale cyan for remaining parts; homodimeric bHLH-Zip transcription factor targets, red for superimposed parts, grey for remaining parts; heterodimeric bHLH-Zip transcription factor targets, red and yellow for superimposed parts, to emphasize the differences in the two chains. The approximate position of the stammer in MITF(wt) is indicated by a dashed line.