Figure 10.
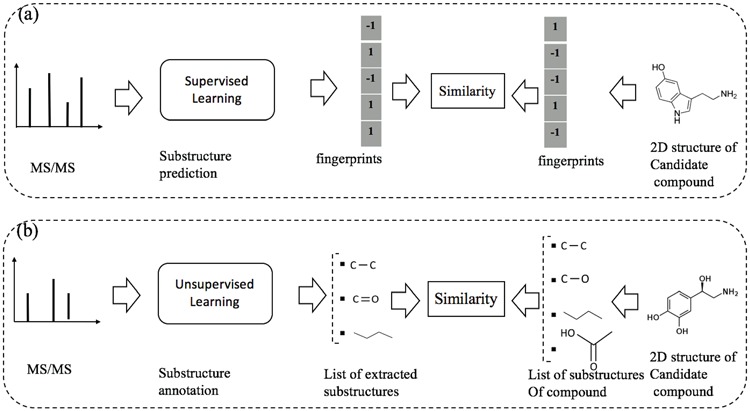
An illustration to clarify the difference between supervised and unsupervised learning for metabolite identification: (a) substructure prediction using supervised learning to map a given MS/MS spectrum to an intermediate representation (e.g. fingerprints), which is subsequently used to retrieve candidate metabolites in the database. (b) substructure annotation using unsupervised learning to extract biochemically relevant substructures with certain confidence from the given spectrum. Then, the similarity between the MS/MS spectrum and a chemical structure of a metabolite is estimated according to their common substructures. Note that the output of supervised learning (e.g. fingerprints) may indicate the presence/absence of all ‘predefined’ substructures whereas that of unsupervised learning may be a list of substructures frequently occurring in the database.
