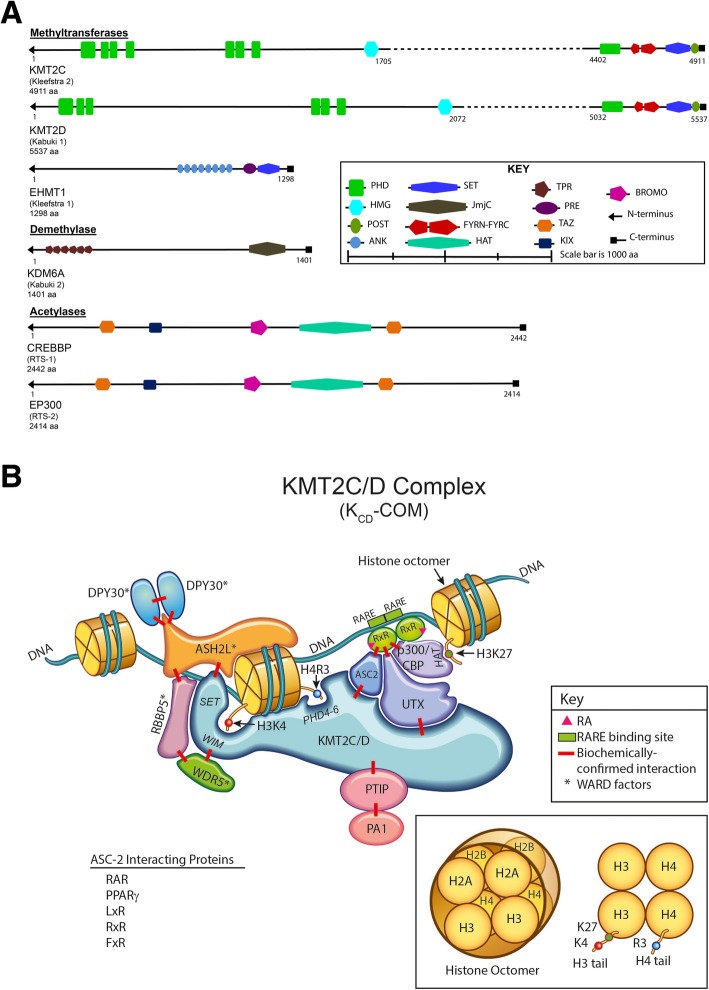
Fig. 1.
a Histone-modifying genes of interest (KMT2C, KMT2D, EHMT1, KDM6A, CRBBP, and EP300) functional domains scale schematic. Amino acids (aa), Plant homeodomain (PHD), high mobility group domain (HMG), F/Y-rich N-terminus domain (FYRN), F/Y-rich C-terminus domain (FYRC), SET domain (SET), post-SET domain (POST), pre-SET domain (PRE), Ankyrin repeat domain (ANK), JmjC domain (JmjC), tetratricopeptide repeat protein domain (TPR), transcriptional adapter zinc binding domain (TAZ), kinase-inducible domain interacting domain (KIX), bromodomain (BROMO), histone acetyltransferase domain (HAT). b Schematic of KCDCOM. COMPASS-like complexes (KCDCOMs, also historically known as ASC2-binding complexes, ASCOMs) bind multiple unique subunits (NCOA6/ASC2, KDM6A/UTX, PTIP, and PAGR1/PA1) and interact with chromatin via histone tail post-translational modifications and DNA binding cofactors. RA retinoic acid. WRAD WDR5 (WD repeat domain 5), RBBP5 (retinoblastoma binding protein 5), ASHL2 (absent, small or homeotic 2-like), and DPY-30 (Dumpy-30)