Fig. 1.
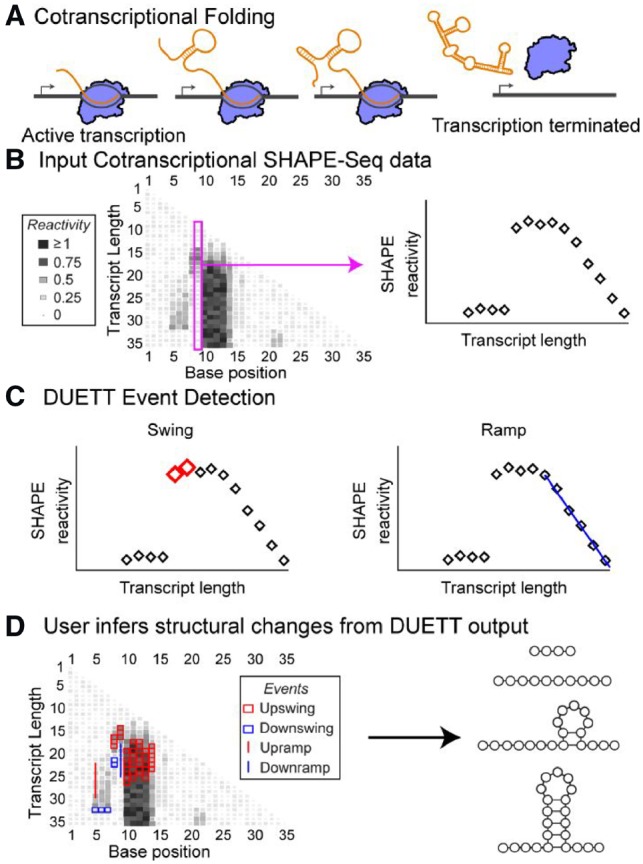
DUETT provides an automated and systematic method to detect cotranscriptional RNA folding events from SHAPE-Seq data. (A) RNA can dynamically alter structure during transcription that affects downstream biological functions. (B) Cotranscriptional SHAPE-Seq probes RNA structural properties during transcription by measuring reactivity patterns for each intermediate length of an RNA. High and low reactivity correspond to unstructured and constrained regions of RNA, respectively. (C) DUETT is a flexible method to identify large and gradual reactivity changes that are indicative of RNA structural transitions that can happen between intermediate RNA lengths. (D) Here DUETT is applied to a mock dataset to identify changes in reactivities in several consecutive nucleotides corresponding to the formation of an RNA loop, which is consistent with the typical ‘low-high-low’ pattern observed for RNA hairpin structures
