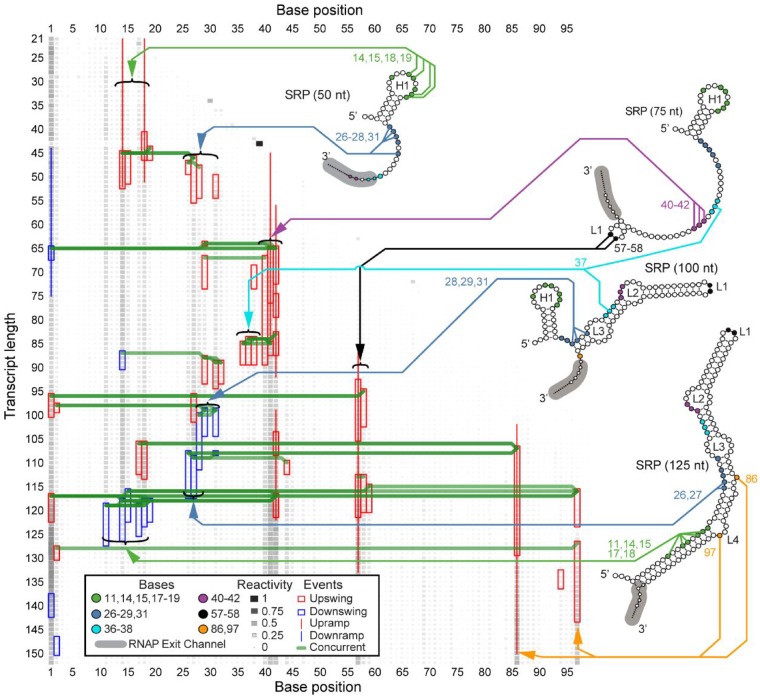
Fig. 2.
DUETT identifies known RNA folding events in the E.coli SRP RNA cotranscriptional SHAPE-Seq reactivity matrices. Four previously proposed intermediate structural conformations of SRP RNA are shown with arrows linking specific color-coded bases to identified reactivity changes. RNA structures are redrawn from Figure 2 of Watters et al. (2016b) with intermediate hairpin H1 and loops L1–L4 labeled, and the RNAP exit channel footprint annotated in gray. DUETT identifies multiple instances of hairpin formation/rearrangement and previously unidentified events. DUETT displays detected swing and ramp events as a colored box and line, respectively, with red and blue denoting reactivity increase and decrease events, respectively. A green line connects concurrent events between different nucleotides. SHAPE reactivity is normalized to lie in between the range 0–1 and shown in grayscale and box area