Figure 6.
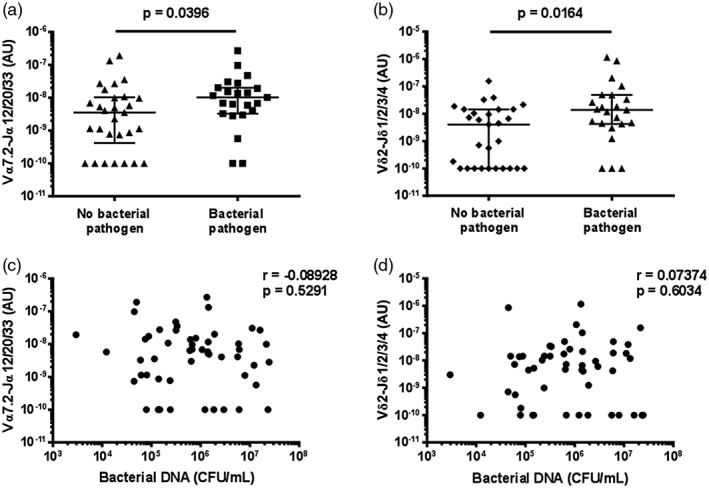
Mucosal‐associated invariant T (MAIT) cell and Vδ2+ T cell abundance in sputum is higher in patients with an identified bacterial pathogen. The abundance of (a) MAIT cells (Vα7.2‐Jα12/20/33 gDNA) and (b) Vδ2+ T cells (Vδ2‐Jδ1/2/3/4 gDNA) in sputum of patients with and without an identified bacterial pathogen. The association between (c) MAIT cell and (d) Vδ2+ T cell abundance in sputum and total bacterial load. MAIT cell and Vδ2+ T cell abundance and total bacterial load were measured by quantitative polymerase chain reaction (qPCR); the absolute copy number of β2 microglobulin (β2M) was calculated with LinRegPCR 23 and the number of MAIT cells (Vα7.2‐Jα12/20/33 gDNA) and Vδ2+ T cells (Vδ2‐Jδ1/2/3/4 gDNA) determined relative to β2M and expressed in arbitrary units. Bacterial pathogens were identified in the diagnostic laboratory by sputum culture, blood culture, PCR on sputum for Legionella spp., or by urinary antigen detection for Streptococcus pneumoniae or L. pneumophila serotype 1 (Supporting information, Table S4). Individual data points (n = 52) representing the mean of technical triplicates of the qPCR assay are shown; in (a) and (b) the median and interquartile ranges are shown. Differences between groups were analysed by Mann–Whitney tests (a,b). The association with total bacterial load was assessed with Spearman correlations (c,d). CFU/ml = colony‐forming units per millilitre.
