Figure 2.
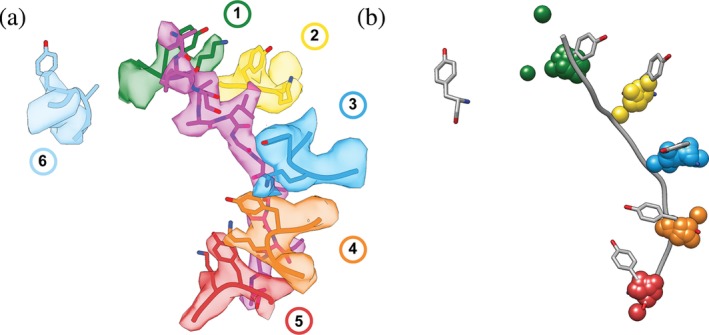
Spiral‐staircase arrangement of pore‐1‐loop residues in contact with substrate. (a) Atomic model (PDB:6EF2) and EM density (EMDB 9440) for the spiral staircase of pore‐1‐loop residues in the 5T state of the 26S proteasome. Shown are the substrate in magenta as well as the conserved Tyr and flanking Lys residues for each protomer numbered by the position in the “spiral staircase,” with Rpt1 in green, Rpt2 in yellow, Rpt6 in blue, Rpt3 in orange, Rpt4 in red, and Rpt5 in cyan. (b) The pore‐1‐loop Tyr's and the substrate polypeptide from the 5T model of the 26S proteasome (56) are displayed in gray and overlaid with 1‐Å centroids indicating the Cα positions for the aromatic “paddle” residues in the pore‐1 loops of substrate‐bound YME1 (6AZ0), AFG3L2 (6NYY), VPS4 (5UIE, 6BMF, 6AP1), the 26S proteasome (1D*‐state (6EF0), 5D‐state (6EF1), 5T‐state (6EF2), 4D‐state (6EF3), ED1‐state (6MSJ), ED2‐state (6MSK)), D2 domains of VAT/p97 (5VCA), D2 domains of NSF (6MDO, 6MDP), D2 domains of Rix7 (6MAT), D2 domains of Hsp104 in the closed (5VJH) and extended states (5VYA), D2 domains of ClpB D2 (ATPγS‐bound double‐WB mutant (5OFO), KC‐1 state (6qs6), KC‐2A state (6qs7), KC‐2B state (6qs8), KC‐3 state (6qs4), WT‐1 (6rn2), WT‐2A (6rn3), WT‐2B (6rn4), conformer1 (6DJU), conformer2 (6DJV), Pre (6OAX), Post (6OAY)), TRIP13 (6F0X), D2 domains of Hsp101 (6E10), and Spastin (6P07). The Cα's for the aromatic pore‐1‐loop residue of protomers 1–5 in each motor were aligned to the proteasomal 5T state as a reference model (6EF2), using Matchmaker in Chimera.75 RMSD values for this alignment ranged from 0.403–1.526 Å. Centroids for the substrate‐disengaged subunit were omitted, as they have the largest variance in position. The two green centroids that do not overlay are from AFG3L2 (6NYY) and 1D* (6EF0), which both have two substrate‐disengaged subunits
