Figure 2.
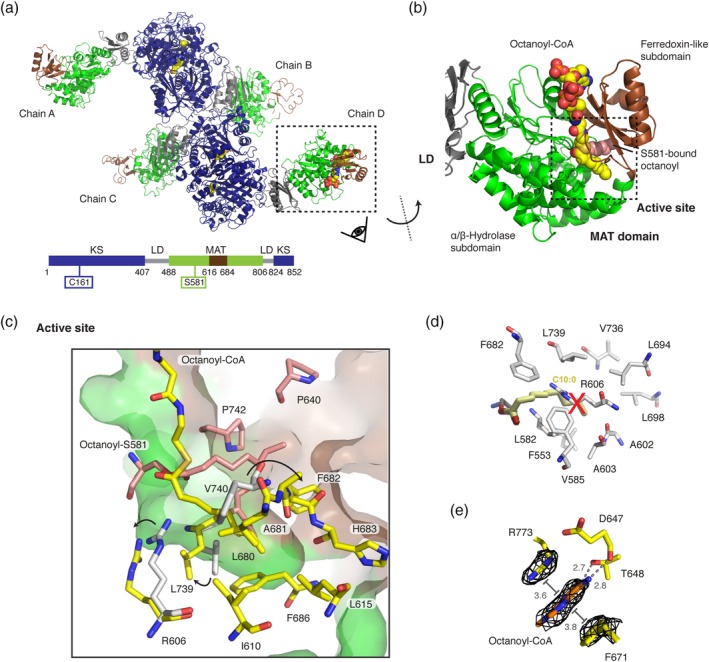
Octanoyl‐loaded MAT domain. (a) Polypeptide chains in the unit cell with bound octanoyl moieties in yellow in sphere representation. Domains and folds are colored as depicted in the attached cartoon. (b) Zoom into the MAT domain in chain D. The active site is embedded in a cleft between the α/β‐hydrolase (green) and the ferredoxin‐like (brown) subdomains. The active site serine (S581) was found in an octanoyl‐bound state with an additional octanoyl‐CoA molecule non‐covalently attached to the active site tunnel (see Figure S2 for FEM38 and Polder maps39). (c) Zoom into the MAT active site. Residues interacting with the serine‐bound octanoyl chain and the octanoyl‐CoA are colored in red and yellow, respectively. Movements of select residues upon binding of an octanoyl moiety in comparison with the human MAT structure (gray) are indicated by arrows. (d) Orientation of a decanoyl chain as reported by Bunkoczi et al.40 Atomic coordinated originate from decanoyl chain computationally modeled into the human MAT variant R606A (PDB code: http://firstglance.jmol.org/fg.htm?mol=2jfd). Postulated interacting residues of the human MAT domain are shown in gray. (e) Binding site of the nucleobase of the CoA moiety at the MAT surface. The adenine is coordinated via hydrogen bonding with residues D647 and T648 and via π‐stacking and π‐cation interactions with residues F671 and R773 were identified. FEM, feature‐enhanced map; MAT, malonyl‐/acetyltransferase
