Figure 5.
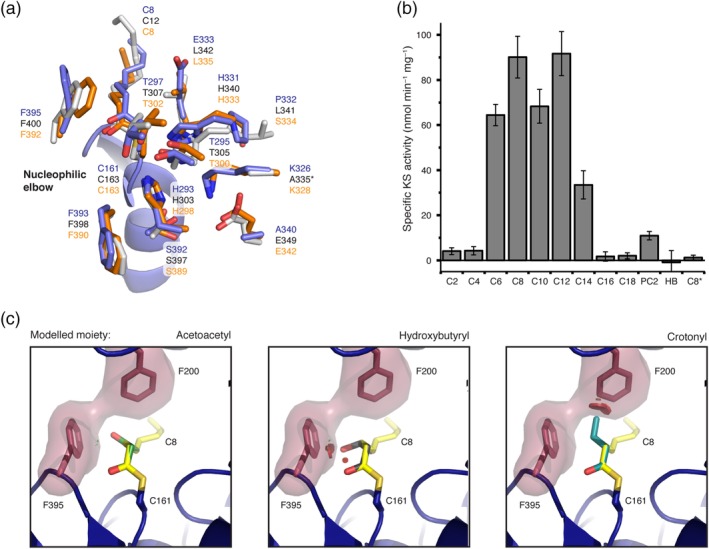
Chain‐length specificity of KS and comparison of the KS domain with FabB and FabF from E. coli. (a) Comparison of important active site residues of the murine Type I KS domain (chain A; blue) with FabB (Orange; PDB code: http://firstglance.jmol.org/fg.htm?mol=2bui) and FabF (gray; PDB code: http://firstglance.jmol.org/fg.htm?mol=2gfy) from E. coli.48, 49 All three proteins were solved in the acyl‐bound state and E. coli proteins were aligned to chain A (blue) by a KS based superposition (BB of residues 1–407). The asterisk indicates that a variant of FabF was crystalized possessing a K335A mutation. (b) Chain‐length specific KS‐mediated transacylation activity. The specific KS‐mediated activity was determined at fixed substrate (500 μM) and holo‐ACP (75 μM) concentrations using the αKGDH‐assay. The asterisk indicates usage of variant KSC161GMATS581A as negative control. Abbreviations refer to acyl‐CoA esters with different chain lengths and PC2 and HB refer to phenylacetyl‐CoA and hydroxybutyryl‐CoA, respectively. (c) Substrate selectivity of the KS domain by gating of the active site through the gatekeeper residue F395 and F200 (both red). The three moieties acetoacetyl (green), hydroxybutyryl (gray), and crotonyl (cyan), common intermediates of fatty acid synthesis, were modeled in place of the bound octanoyl moiety. Clashes are highlighted by red bumps. ACP, acyl carrier protein; BB, backbone atoms; KS, ketosynthase
