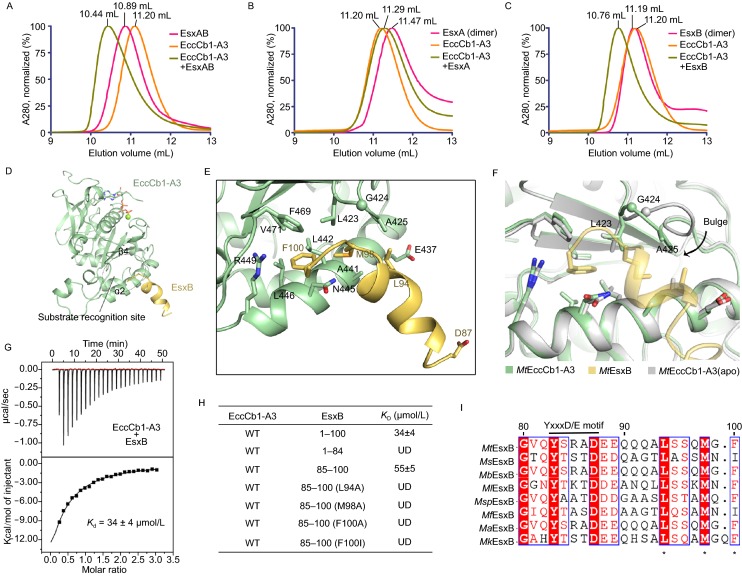
Figure 3.
The C-terminal peptide ofMtEsxB interacts withMtEccCb1-ATPase3. (A–C) Gel filtration analysis of interactions between Esx proteins and MtEccCb1-ATPase3 performed on a Superdex 75 column. The peak volumes are indicated on the top. (A) MtEsxAB binds to MtEccCb1-ATPase3 inducing a shift in elution volume. (B) MtEsxA alone does not bind to MtEccCb1-ATPase3, thus there is no change in elution volume. (C) MtEsxB binds to MtEccCb1-ATPase3, inducing a similar shift in elution volume as observed in (A). (D) Overall structure of MtEccCb1-ATPase3 (palegreen) complexed with MtEsxB (yelloworange). The substrate binding site is marked with a dashed circle. (E) Detailed interactions between MtEsxB and MtEccCb1-ATPase3. Interacting residues and Asp87 in Yxxx[D/E] motif are shown as sticks. (F) MtEccCb1-ATPase3 in complex with MtEsxB is superimposed onto MtEccCb1-ATPase3 (apo). The bulge in the loop moves closer to MtEsxB to enhance substrate binding. (G) ITC assay shows the binding affinity of MtEsxB to MtEccCb1-ATPase3. The data were representative of at least three repetitions. (H) The dissociation constant, Kd, is based on the ITC studies of MtEsxB and its truncations or peptides, to MtEccCb1-ATPase3. The data were representative of at least three repetitions. WT, wild type; UD, data was undetermined. (I) Sequence alignment of EsxB from different Mycobacterium species including Mycobacterium tuberculosis (Mt), Mycobacterium smegmatis (Ms), Mycobacterium bovis (Mb), Mycobacterium leprae (Ml), Mycobacterium sp. (Msp), Mycobacterium flavescens (Mf), Mycobacterium africanum (Ma) and Mycobacterium kyorinense (Mk). The recognition residues are marked with stars. The Yxxx[D/E] motif is also labeled on top of sequences