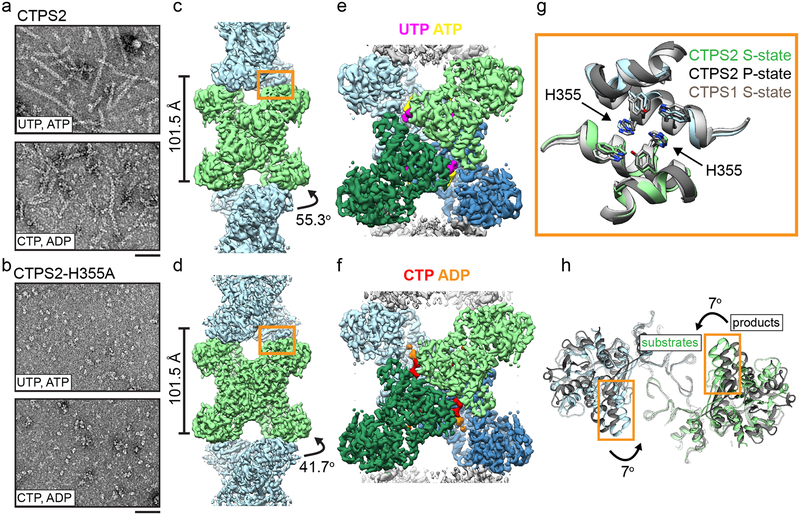
Fig. 1: CTPS2 forms distinct S and P-state filaments using the same filament interface.
(a,b) Negative stain EM images of CTPS2 wild-type (a) or CTPS2-H355A non-polymerizing mutant (b) in the presence of substrates or products. Scale bars are 50 nm. (c,d) Initial cryo-EM reconstructions of S-state (c, 4Å resolution) and P-state (d, 3.6 Å resolution) CTPS2 filaments showing differing helical symmetry, colored by tetramer. Helical rise and rotation are indicated. (e,f) High-resolution reconstructions of S-state (e, 3.5Å resolution) and P-state (f, 3.1 Å resolution) CTPS2 filaments focused on a single tetramer, colored by protomer. Nucleotides are colored as indicated. (g) The filament interface (orange box in c,d) is conserved in the CTPS2 S-state (color), CTPS2 P-state (dark grey), and CTPS1 S-state (light grey) structures. Position of conserved residue H355 is indicated. (h) View down the helical axis comparing the positions of the filament contacts (orange box) in the S-state (color) and P-state (grey) filaments.