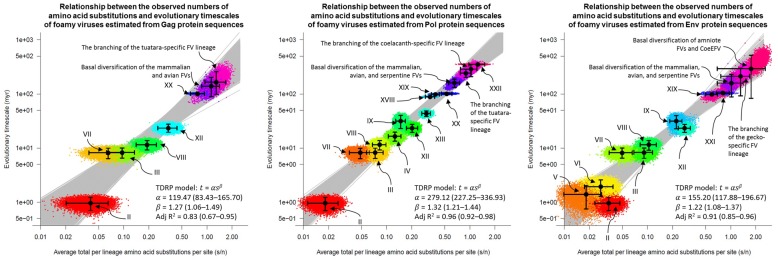
Figure 3.
TDRP models estimated from the Gag (left), Pol (middle), and Env (right) proteins. The models were calibrated using average node-to-tip distances (estimates) and their corresponding evolutionary timescales ( estimates) inferred under the FV-host co-speciation assumption (Switzer et al. 2005; Katzourakis et al. 2009, 2014; Ghersi et al. 2015). Those derived from the co-speciation events are labelled with Roman numerals, referring to nodes in Fig. 2. Solid black dots are median estimates, and the associated 95 per cent HPD intervals are indicated by error bars. A total of 7,500 TDRP models were fitted to the posterior distributions of the and estimates (see Materials and Methods). The median model parameter values, adjusted R2 scores, and corresponding 95 per cent HPDs (in the parentheses) are shown in the bottom right. The models were extrapolated to infer the dates of various other events as labelled. See Supplementary Table S3 for the values of and estimates.