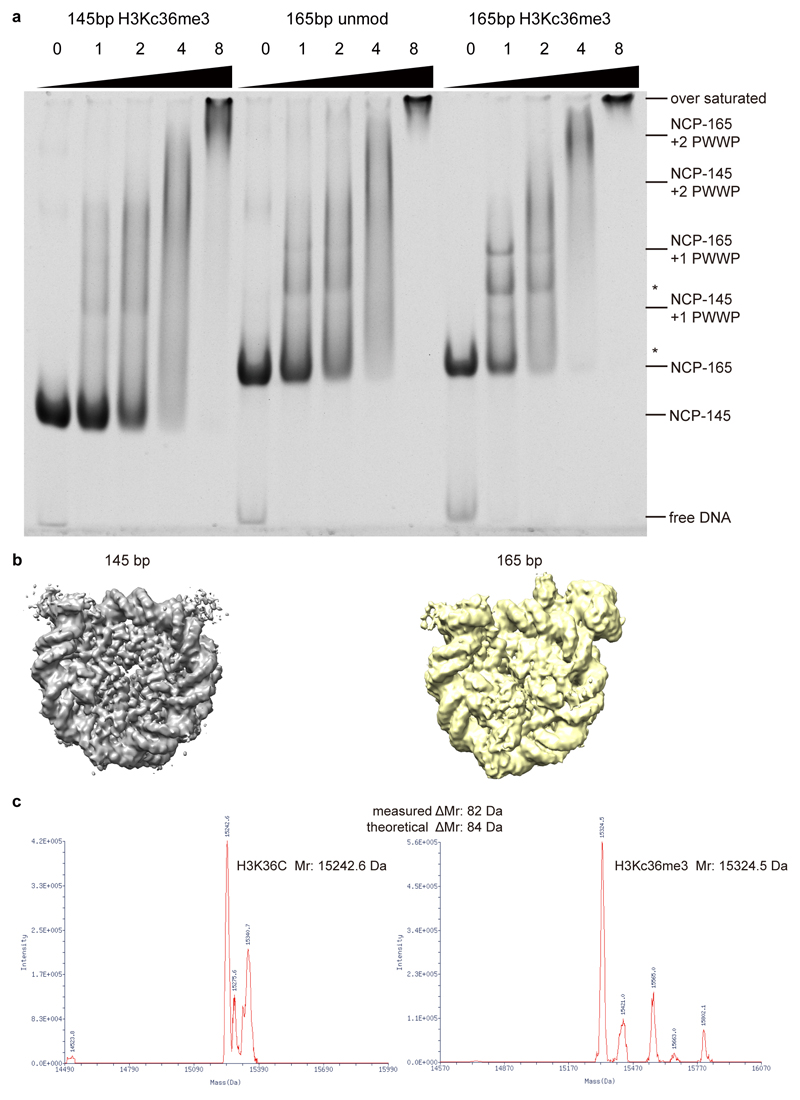
Extended Data Fig. 1. Binding of LEDGF to H3KC36me3-modified nucleosome.
a. EMSA reveals that full-length LEDGF preferentially binds to a H3KC36me3-modified nucleosome with longer (165 bp) DNA. Molar ratio of full-length LEDGF to indicated nucleosomes are shown on the top of each lane. Bands correspond to each component and complexes are labeled on the right. Bands of degraded LEDGF-bound nucleosomes are denoted with *. For gel source data, see Source Data Extended Data Fig. 1.
b. Reconstructed EM density maps of 145bp and 165bp H3KC36me3-modified nucleosome with LEDGF. Note that the presence of the extra DNA in the latter complex leads to a defined additional density for the PWWP domain.
c. Mass spectrometry measurement of the H3KC36me3 modified histone H3. Left: molecular weight measurement of H3K36C mutant. Right: molecular weight measurement of H3KC36me3 modified H3.